Listed below are N-terminal peptides that were captured and enriched from the surface of a HEK293T cell line expressing subtiligase-TM.
RPN1_HUMAN
P04843 - Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1
FUNCTION: Essential subunit of the N-oligosaccharyl transferase (OST) complex which catalyzes the transfer of a high mannose oligosaccharide from a lipid-linked oligosaccharide donor to an asparagine residue within an Asn-X-Ser/Thr consensus motif in nascent polypeptide chains. {ECO:0000250|UniProtKB:E2RQ08, ECO:0000250|UniProtKB:Q9GMB0, ECO:0000305}.
SUBCELLULAR LOCATION: Endoplasmic reticulum {ECO:0000250|UniProtKB:E2RQ08, ECO:0000250|UniProtKB:Q9GMB0}. Endoplasmic reticulum membrane; Single-pass type I membrane protein {ECO:0000305}. Melanosome. Note=Identified by mass spectrometry in melanosome fractions from stage I to stage IV.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TBA1B_HUMAN FUNCTION: Tubulin is the major constituent of microtubules. It
binds two moles of GTP, one at an exchangeable site on the beta
chain and one at a non-exchangeable site on the alpha chain.
SUBCELLULAR LOCATION: Cytoplasm, cytoskeleton.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CADH2_HUMAN FUNCTION: Cadherins are calcium-dependent cell adhesion proteins.
They preferentially interact with themselves in a homophilic
manner in connecting cells; cadherins may thus contribute to the
sorting of heterogeneous cell types. Acts as a regulator of neural
stem cells quiescence by mediating anchorage of neural stem cells
to ependymocytes in the adult subependymal zone: upon cleavage by
MMP24, CDH2-mediated anchorage is affected, leading to modulate
neural stem cell quiescence. CDH2 may be involved in neuronal
recognition mechanism. In hippocampal neurons, may regulate
dendritic spine density (By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Cell membrane
{ECO:0000250|UniProtKB:P15116}; Single-pass type I membrane
protein {ECO:0000255}. Cell membrane, sarcolemma
{ECO:0000250|UniProtKB:P15116}. Cell junction
{ECO:0000269|PubMed:28169360}. Cell surface
{ECO:0000250|UniProtKB:P15116}. Note=Colocalizes with TMEM65 at
the intercalated disk in cardiomyocytes. Colocalizes with OBSCN at
the intercalated disk and at sarcolemma in cardiomyocytes.
{ECO:0000250|UniProtKB:P15116}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: GRP78_HUMAN FUNCTION: Probably plays a role in facilitating the assembly of
multimeric protein complexes inside the endoplasmic reticulum.
Involved in the correct folding of proteins and degradation of
misfolded proteins via its interaction with DNAJC10, probably to
facilitate the release of DNAJC10 from its substrate.
{ECO:0000269|PubMed:2294010, ECO:0000269|PubMed:23769672,
ECO:0000269|PubMed:23990668}.
SUBCELLULAR LOCATION: Endoplasmic reticulum lumen
{ECO:0000269|PubMed:21080038, ECO:0000269|PubMed:21289099,
ECO:0000269|PubMed:23990668}. Melanosome. Cytoplasm {ECO:0000250}.
Note=Identified by mass spectrometry in melanosome fractions from
stage I to stage IV.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CALX_HUMAN FUNCTION: Calcium-binding protein that interacts with newly
synthesized glycoproteins in the endoplasmic reticulum. It may act
in assisting protein assembly and/or in the retention within the
ER of unassembled protein subunits. It seems to play a major role
in the quality control apparatus of the ER by the retention of
incorrectly folded proteins. Associated with partial T-cell
antigen receptor complexes that escape the ER of immature
thymocytes, it may function as a signaling complex regulating
thymocyte maturation. Additionally it may play a role in receptor-
mediated endocytosis at the synapse.
SUBCELLULAR LOCATION: Endoplasmic reticulum membrane
{ECO:0000269|PubMed:22314232}; Single-pass type I membrane protein
{ECO:0000255}. Endoplasmic reticulum
{ECO:0000269|PubMed:22314232}. Melanosome
{ECO:0000269|PubMed:12643545, ECO:0000269|PubMed:17081065}.
Note=Identified by mass spectrometry in melanosome fractions from
stage I to stage IV (PubMed:12643545, PubMed:17081065). The
palmitoylated form preferentially localizes to the perinuclear
rough ER (PubMed:22314232). {ECO:0000269|PubMed:12643545,
ECO:0000269|PubMed:17081065, ECO:0000269|PubMed:22314232}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: ENPL_HUMAN FUNCTION: Molecular chaperone that functions in the processing and
transport of secreted proteins. When associated with CNPY3,
required for proper folding of Toll-like receptors (By
similarity). Functions in endoplasmic reticulum associated
degradation (ERAD). Has ATPase activity. {ECO:0000250,
ECO:0000269|PubMed:18264092}.
SUBCELLULAR LOCATION: Endoplasmic reticulum lumen. Melanosome.
Note=Identified by mass spectrometry in melanosome fractions from
stage I to stage IV.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: 1A02_HUMAN FUNCTION: Involved in the presentation of foreign antigens to the
immune system.
SUBCELLULAR LOCATION: Membrane; Single-pass type I membrane
protein.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CD99_HUMAN FUNCTION: Involved in T-cell adhesion processes and in spontaneous
rosette formation with erythrocytes. Plays a role in a late step
of leukocyte extravasation helping leukocytes to overcome the
endothelial basement membrane. Acts at the same site as, but
independently of, PECAM1. Involved in T-cell adhesion processes
(By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Membrane {ECO:0000305}; Single-pass type I
membrane protein {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: PTPRF_HUMAN FUNCTION: Possible cell adhesion receptor. It possesses an
intrinsic protein tyrosine phosphatase activity (PTPase) and
dephosphorylates EPHA2 regulating its activity.
FUNCTION: The first PTPase domain has enzymatic activity, while
the second one seems to affect the substrate specificity of the
first one.
SUBCELLULAR LOCATION: Membrane; Single-pass type I membrane
protein.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: EF1A1_HUMAN FUNCTION: This protein promotes the GTP-dependent binding of
aminoacyl-tRNA to the A-site of ribosomes during protein
biosynthesis. With PARP1 and TXK, forms a complex that acts as a T
helper 1 (Th1) cell-specific transcription factor and binds the
promoter of IFN-gamma to directly regulate its transcription, and
is thus involved importantly in Th1 cytokine production.
{ECO:0000269|PubMed:17177976}.
SUBCELLULAR LOCATION: Cytoplasm {ECO:0000269|PubMed:19158340,
ECO:0000269|PubMed:8650580}. Nucleus {ECO:0000269|PubMed:17177976,
ECO:0000269|PubMed:8650580}. Nucleus, nucleolus
{ECO:0000269|PubMed:8650580}. Cell membrane
{ECO:0000269|PubMed:26497934}. Note=Colocalizes with DLC1 at
actin-rich regions in the cell periphery (PubMed:19158340).
Translocates together with ZPR1 from the cytoplasm to the nucleus
and nucleolus after treatment with mitogens (PubMed:8650580).
Localization at the cell membrane depends on EEF1A1
phosphorylation status and the presence of PPP1R16B
(PubMed:26497934). {ECO:0000269|PubMed:19158340,
ECO:0000269|PubMed:26497934, ECO:0000269|PubMed:8650580}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: BASI_HUMAN FUNCTION: Plays an important role in targeting the monocarboxylate
transporters SLC16A1, SLC16A3 and SLC16A8 to the plasma membrane.
Plays pivotal roles in spermatogenesis, embryo implantation,
neural network formation and tumor progression. Stimulates
adjacent fibroblasts to produce matrix metalloproteinases (MMPS).
Seems to be a receptor for oligomannosidic glycans. In vitro,
promotes outgrowth of astrocytic processes.
{ECO:0000269|PubMed:17127621}.
SUBCELLULAR LOCATION: Cell membrane {ECO:0000269|PubMed:15946952,
ECO:0000269|PubMed:17127621, ECO:0000269|PubMed:21536654}; Single-
pass type I membrane protein {ECO:0000303|PubMed:15946952}.
Melanosome {ECO:0000269|PubMed:17081065}. Note=Identified by mass
spectrometry in melanosome fractions from stage I to stage IV. In
spermatozoa, localized on the principal piece of caput and in the
middle piece during transit in the corpus and cauda epididymides
(By similarity). {ECO:0000250|UniProtKB:P18572,
ECO:0000269|PubMed:17081065}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: ATPB_HUMAN FUNCTION: Mitochondrial membrane ATP synthase (F(1)F(0) ATP
synthase or Complex V) produces ATP from ADP in the presence of a
proton gradient across the membrane which is generated by electron
transport complexes of the respiratory chain. F-type ATPases
consist of two structural domains, F(1) - containing the
extramembraneous catalytic core, and F(0) - containing the
membrane proton channel, linked together by a central stalk and a
peripheral stalk. During catalysis, ATP synthesis in the catalytic
domain of F(1) is coupled via a rotary mechanism of the central
stalk subunits to proton translocation. Subunits alpha and beta
form the catalytic core in F(1). Rotation of the central stalk
against the surrounding alpha(3)beta(3) subunits leads to
hydrolysis of ATP in three separate catalytic sites on the beta
subunits.
SUBCELLULAR LOCATION: Mitochondrion. Mitochondrion inner membrane.
Note=Peripheral membrane protein.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: HYOU1_HUMAN FUNCTION: Has a pivotal role in cytoprotective cellular mechanisms
triggered by oxygen deprivation. May play a role as a molecular
chaperone and participate in protein folding.
{ECO:0000269|PubMed:10037731}.
SUBCELLULAR LOCATION: Endoplasmic reticulum lumen.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: AAAT_HUMAN FUNCTION: Sodium-dependent amino acids transporter that has a
broad substrate specificity, with a preference for zwitterionic
amino acids. It accepts as substrates all neutral amino acids,
including glutamine, asparagine, and branched-chain and aromatic
amino acids, and excludes methylated, anionic, and cationic amino
acids (PubMed:8702519). Through binding of the fusogenic protein
syncytin-1/ERVW-1 may mediate trophoblasts syncytialization, the
spontaneous fusion of their plasma membranes, an essential process
in placental development (PubMed:10708449, PubMed:23492904).
{ECO:0000269|PubMed:10708449, ECO:0000269|PubMed:23492904,
ECO:0000269|PubMed:8702519}.
FUNCTION: (Microbial infection) Acts as a cell surface receptor
for feline endogenous virus RD114, baboon M7 endogenous virus and
type D simian retroviruses. {ECO:0000269|PubMed:10051606,
ECO:0000269|PubMed:10196349}.
SUBCELLULAR LOCATION: Cell membrane {ECO:0000269|PubMed:8702519};
Multi-pass membrane protein {ECO:0000305}. Melanosome.
Note=Identified by mass spectrometry in melanosome fractions from
stage I to stage IV.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: DSG2_HUMAN FUNCTION: Component of intercellular desmosome junctions. Involved
in the interaction of plaque proteins and intermediate filaments
mediating cell-cell adhesion.
SUBCELLULAR LOCATION: Cell membrane; Single-pass type I membrane
protein. Cell junction, desmosome.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TM9S2_HUMAN FUNCTION: In the intracellular compartments, may function as a
channel or small molecule transporter.
SUBCELLULAR LOCATION: Endosome membrane {ECO:0000305}; Multi-pass
membrane protein {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TXND5_HUMAN FUNCTION: Possesses thioredoxin activity. Has been shown to reduce
insulin disulfide bonds. Also complements protein disulfide-
isomerase deficiency in yeast (By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Endoplasmic reticulum lumen
{ECO:0000255|PROSITE-ProRule:PRU10138}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: AT2A2_HUMAN FUNCTION: This magnesium-dependent enzyme catalyzes the hydrolysis
of ATP coupled with the translocation of calcium from the cytosol
to the sarcoplasmic reticulum lumen. Isoform 2 is involved in the
regulation of the contraction/relaxation cycle (PubMed:16402920).
Acts as a regulator of TNFSF11-mediated Ca(2+) signaling pathways
via its interaction with TMEM64 which is critical for the TNFSF11-
induced CREB1 activation and mitochondrial ROS generation
necessary for proper osteoclast generation. Association between
TMEM64 and SERCA2 in the ER leads to cytosolic Ca (2+) spiking for
activation of NFATC1 and production of mitochondrial ROS, thereby
triggering Ca (2+) signaling cascades that promote osteoclast
differentiation and activation (By similarity).
{ECO:0000250|UniProtKB:O55143, ECO:0000269|PubMed:16402920}.
SUBCELLULAR LOCATION: Endoplasmic reticulum membrane
{ECO:0000250|UniProtKB:O55143}; Multi-pass membrane protein
{ECO:0000255}. Sarcoplasmic reticulum membrane
{ECO:0000250|UniProtKB:O55143}; Multi-pass membrane protein
{ECO:0000255}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: RPN2_HUMAN FUNCTION: Essential subunit of the N-oligosaccharyl transferase
(OST) complex which catalyzes the transfer of a high mannose
oligosaccharide from a lipid-linked oligosaccharide donor to an
asparagine residue within an Asn-X-Ser/Thr consensus motif in
nascent polypeptide chains. {ECO:0000250|UniProtKB:F1PCT7,
ECO:0000305}.
SUBCELLULAR LOCATION: Endoplasmic reticulum
{ECO:0000250|UniProtKB:F1PCT7}. Endoplasmic reticulum membrane;
Multi-pass membrane protein {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CD166_HUMAN FUNCTION: Cell adhesion molecule that mediates both heterotypic
cell-cell contacts via its interaction with CD6, as well as
homotypic cell-cell contacts (PubMed:7760007, PubMed:15496415,
PubMed:15048703, PubMed:16352806, PubMed:23169771,
PubMed:24945728). Promotes T-cell activation and proliferation via
its interactions with CD6 (PubMed:15048703, PubMed:16352806,
PubMed:24945728). Contributes to the formation and maturation of
the immunological synapse via its interactions with CD6
(PubMed:15294938, PubMed:16352806). Mediates homotypic
interactions with cells that express ALCAM (PubMed:15496415,
PubMed:16352806). Required for normal hematopoietic stem cell
engraftment in the bone marrow (PubMed:24740813). Mediates
attachment of dendritic cells onto endothelial cells via homotypic
interaction (PubMed:23169771). Inhibits endothelial cell migration
and promotes endothelial tube formation via homotypic interactions
(PubMed:15496415, PubMed:23169771). Required for normal
organization of the lymph vessel network. Required for normal
hematopoietic stem cell engraftment in the bone marrow. Plays a
role in hematopoiesis; required for normal numbers of
hematopoietic stem cells in bone marrow. Promotes in vitro
osteoblast proliferation and differentiation (By similarity).
Promotes neurite extension, axon growth and axon guidance; axons
grow preferentially on surfaces that contain ALCAM. Mediates
outgrowth and pathfinding for retinal ganglion cell axons (By
similarity). {ECO:0000250|UniProtKB:P42292,
ECO:0000269|PubMed:15048703, ECO:0000269|PubMed:15294938,
ECO:0000269|PubMed:15496415, ECO:0000269|PubMed:16352806,
ECO:0000269|PubMed:24945728, ECO:0000269|PubMed:7760007}.
FUNCTION: Isoform 3: Inhibits activities of membrane-bound
isoforms by competing for the same interaction partners. Inhibits
cell attachment via homotypic interactions. Promotes endothelial
cell migration. Inhibits endothelial cell tube formation.
{ECO:0000269|PubMed:15496415}.
SUBCELLULAR LOCATION: Cell membrane {ECO:0000269|PubMed:15048703,
ECO:0000269|PubMed:15294938, ECO:0000269|PubMed:16352806,
ECO:0000269|PubMed:23169771, ECO:0000269|PubMed:24740813,
ECO:0000269|PubMed:24945728, ECO:0000269|PubMed:7760007}; Single-
pass type I membrane protein {ECO:0000305}. Cell projection, axon
{ECO:0000250|UniProtKB:Q61490}. Cell projection, dendrite
{ECO:0000250|UniProtKB:Q61490}. Note=Detected at the immunological
synapse, i.e, at the contact zone between antigen-presenting
dendritic cells and T-cells (PubMed:15294938, PubMed:16352806).
Colocalizes with CD6 and the TCR/CD3 complex at the immunological
synapse (PubMed:15294938). {ECO:0000269|PubMed:15294938,
ECO:0000269|PubMed:16352806}.
SUBCELLULAR LOCATION: Isoform 3: Secreted
{ECO:0000269|PubMed:15496415}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: GLU2B_HUMAN FUNCTION: Regulatory subunit of glucosidase II.
{ECO:0000269|PubMed:10929008}.
SUBCELLULAR LOCATION: Endoplasmic reticulum {ECO:0000255|PROSITE-
ProRule:PRU10138}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TGO1_HUMAN FUNCTION: Plays a role in the transport of cargos that are too
large to fit into COPII-coated vesicles and require specific
mechanisms to be incorporated into membrane-bound carriers and
exported from the endoplasmic reticulum. This protein is required
for collagen VII (COL7A1) secretion by loading COL7A1 into
transport carriers. It may participate in cargo loading of COL7A1
at endoplasmic reticulum exit sites by binding to COPII coat
subunits Sec23/24 and guiding SH3-bound COL7A1 into a growing
carrier. Does not play a role in global protein secretion and is
apparently specific to COL7A1 cargo loading. However, it may
participate in secretion of other proteins in cells that do not
secrete COL7A1. It is also specifically required for the secretion
of lipoproteins by participating in their export from the
endoplasmic reticulum (PubMed:27138255).
{ECO:0000269|PubMed:19269366, ECO:0000269|PubMed:27138255}.
SUBCELLULAR LOCATION: Endoplasmic reticulum membrane
{ECO:0000269|PubMed:19269366, ECO:0000269|PubMed:21525241};
Single-pass membrane protein {ECO:0000269|PubMed:19269366}.
Note=Localizes at endoplasmic reticulum exit sites. After loading
of COL7A1 into transport carriers, it is not incorporated into
COPII carriers and remains in the endoplasmic reticulum membrane.
{ECO:0000269|PubMed:19269366, ECO:0000269|PubMed:21525241}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TR10B_HUMAN FUNCTION: Receptor for the cytotoxic ligand TNFSF10/TRAIL
(PubMed:10549288). The adapter molecule FADD recruits caspase-8 to
the activated receptor. The resulting death-inducing signaling
complex (DISC) performs caspase-8 proteolytic activation which
initiates the subsequent cascade of caspases (aspartate-specific
cysteine proteases) mediating apoptosis. Promotes the activation
of NF-kappa-B. Essential for ER stress-induced apoptosis.
{ECO:0000269|PubMed:10542098, ECO:0000269|PubMed:10549288,
ECO:0000269|PubMed:15322075}.
SUBCELLULAR LOCATION: Membrane; Single-pass type I membrane
protein.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: 4F2_HUMAN FUNCTION: Required for the function of light chain amino-acid
transporters. Involved in sodium-independent, high-affinity
transport of large neutral amino acids such as phenylalanine,
tyrosine, leucine, arginine and tryptophan. Involved in guiding
and targeting of LAT1 and LAT2 to the plasma membrane. When
associated with SLC7A6 or SLC7A7 acts as an arginine/glutamine
exchanger, following an antiport mechanism for amino acid
transport, influencing arginine release in exchange for
extracellular amino acids. Plays a role in nitric oxide synthesis
in human umbilical vein endothelial cells (HUVECs) via transport
of L-arginine. Required for normal and neoplastic cell growth.
When associated with SLC7A5/LAT1, is also involved in the
transport of L-DOPA across the blood-brain barrier, and that of
thyroid hormones triiodothyronine (T3) and thyroxine (T4) across
the cell membrane in tissues such as placenta. Involved in the
uptake of methylmercury (MeHg) when administered as the L-cysteine
or D,L-homocysteine complexes, and hence plays a role in metal ion
homeostasis and toxicity. When associated with SLC7A5 or SLC7A8,
involved in the cellular activity of small molecular weight
nitrosothiols, via the stereoselective transport of L-
nitrosocysteine (L-CNSO) across the transmembrane. Together with
ICAM1, regulates the transport activity LAT2 in polarized
intestinal cells, by generating and delivering intracellular
signals. When associated with SLC7A5, plays an important role in
transporting L-leucine from the circulating blood to the retina
across the inner blood-retinal barrier.
{ECO:0000269|PubMed:10903140, ECO:0000269|PubMed:11311135,
ECO:0000269|PubMed:11389679, ECO:0000269|PubMed:11557028,
ECO:0000269|PubMed:11564694, ECO:0000269|PubMed:11742812,
ECO:0000269|PubMed:12117417, ECO:0000269|PubMed:12225859,
ECO:0000269|PubMed:12716892, ECO:0000269|PubMed:14603368,
ECO:0000269|PubMed:15769744, ECO:0000269|PubMed:15980244,
ECO:0000269|PubMed:9751058, ECO:0000269|PubMed:9829974,
ECO:0000269|PubMed:9878049}.
SUBCELLULAR LOCATION: Apical cell membrane; Single-pass type II
membrane protein. Melanosome. Note=Identified by mass spectrometry
in melanosome fractions from stage I to stage IV. Localized to the
plasma membrane when associated with SLC7A5 or SLC7A8. Localized
to the placental apical membrane. Located selectively at cell-cell
adhesion sites (By similarity). Colocalized with SLC7A8/LAT2 at
the basolateral membrane of kidney proximal tubules and small
intestine epithelia. Expressed in both luminal and abluminal
membranes of brain capillary endothelial cells (By similarity).
{ECO:0000250}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: ITB1_HUMAN FUNCTION: Integrins alpha-1/beta-1, alpha-2/beta-1, alpha-10/beta-
1 and alpha-11/beta-1 are receptors for collagen. Integrins alpha-
1/beta-1 and alpha-2/beta-2 recognize the proline-hydroxylated
sequence G-F-P-G-E-R in collagen. Integrins alpha-2/beta-1, alpha-
3/beta-1, alpha-4/beta-1, alpha-5/beta-1, alpha-8/beta-1, alpha-
10/beta-1, alpha-11/beta-1 and alpha-V/beta-1 are receptors for
fibronectin. Alpha-4/beta-1 recognizes one or more domains within
the alternatively spliced CS-1 and CS-5 regions of fibronectin.
Integrin alpha-5/beta-1 is a receptor for fibrinogen. Integrin
alpha-1/beta-1, alpha-2/beta-1, alpha-6/beta-1 and alpha-7/beta-1
are receptors for lamimin. Integrin alpha-4/beta-1 is a receptor
for VCAM1. It recognizes the sequence Q-I-D-S in VCAM1. Integrin
alpha-9/beta-1 is a receptor for VCAM1, cytotactin and
osteopontin. It recognizes the sequence A-E-I-D-G-I-E-L in
cytotactin. Integrin alpha-3/beta-1 is a receptor for epiligrin,
thrombospondin and CSPG4. Alpha-3/beta-1 may mediate with LGALS3
the stimulation by CSPG4 of endothelial cells migration. Integrin
alpha-V/beta-1 is a receptor for vitronectin. Beta-1 integrins
recognize the sequence R-G-D in a wide array of ligands. Isoform 2
interferes with isoform 1 resulting in a dominant negative effect
on cell adhesion and migration (in vitro). When associated with
alpha-7/beta-1 integrin, regulates cell adhesion and laminin
matrix deposition. Involved in promoting endothelial cell motility
and angiogenesis. Involved in osteoblast compaction through the
fibronectin fibrillogenesis cell-mediated matrix assembly process
and the formation of mineralized bone nodules. May be involved in
up-regulation of the activity of kinases such as PKC via binding
to KRT1. Together with KRT1 and RACK1, serves as a platform for
SRC activation or inactivation. Plays a mechanistic adhesive role
during telophase, required for the successful completion of
cytokinesis. Integrin alpha-3/beta-1 provides a docking site for
FAP (seprase) at invadopodia plasma membranes in a collagen-
dependent manner and hence may participate in the adhesion,
formation of invadopodia and matrix degradation processes,
promoting cell invasion. ITGA4:ITGB1 binds to fractalkine (CX3CL1)
and may act as its coreceptor in CX3CR1-dependent fractalkine
signaling (PubMed:23125415, PubMed:24789099). ITGA4:ITGB1 and
ITGA5:ITGB1 bind to PLA2G2A via a site (site 2) which is distinct
from the classical ligand-binding site (site 1) and this induces
integrin conformational changes and enhanced ligand binding to
site 1 (PubMed:18635536, PubMed:25398877). ITGA5:ITGB1 acts as a
receptor for fibrillin-1 (FBN1) and mediates R-G-D-dependent cell
adhesion to FBN1 (PubMed:12807887, PubMed:17158881).
{ECO:0000269|PubMed:10455171, ECO:0000269|PubMed:12473654,
ECO:0000269|PubMed:12807887, ECO:0000269|PubMed:16256741,
ECO:0000269|PubMed:17158881, ECO:0000269|PubMed:18635536,
ECO:0000269|PubMed:18804435, ECO:0000269|PubMed:19064666,
ECO:0000269|PubMed:21768292, ECO:0000269|PubMed:23125415,
ECO:0000269|PubMed:24789099, ECO:0000269|PubMed:25398877,
ECO:0000269|PubMed:7523423}.
FUNCTION: Isoform 5: Isoform 5 displaces isoform 1 in striated
muscles. {ECO:0000250}.
FUNCTION: (Microbial infection) Integrin ITGA2:ITGB1 acts as a
receptor for human echoviruses 1 and 8 (PubMed:8411387). Acts as a
receptor for cytomegalovirus/HHV-5 (PubMed:20660204). Acts as a
receptor for Epstein-Barr virus/HHV-4 (PubMed:17945327). Integrin
ITGA5:ITGB1 acts as a receptor for human parvovirus B19
(PubMed:12907437). Integrin ITGA2:ITGB1 acts as a receptor for
human rotavirus (PubMed:12941907). Acts as a receptor for
mammalian reovirus (PubMed:16501085). In case of HIV-1 infection,
integrin ITGA5:ITGB1 binding to extracellular viral Tat protein
seems to enhance angiogenesis in Kaposi's sarcoma lesions
(PubMed:10397733). {ECO:0000269|PubMed:10397733,
ECO:0000269|PubMed:12907437, ECO:0000269|PubMed:12941907,
ECO:0000269|PubMed:16501085, ECO:0000269|PubMed:17945327,
ECO:0000269|PubMed:20660204, ECO:0000269|PubMed:8411387}.
SUBCELLULAR LOCATION: Cell membrane {ECO:0000303|PubMed:10455171};
Single-pass type I membrane protein {ECO:0000255}. Cell
projection, invadopodium membrane {ECO:0000269|PubMed:10455171};
Single-pass type I membrane protein {ECO:0000255}. Cell
projection, ruffle membrane {ECO:0000303|PubMed:10455171}; Single-
pass type I membrane protein {ECO:0000255}. Recycling endosome
{ECO:0000269|PubMed:16256741}. Melanosome
{ECO:0000269|PubMed:17081065}. Cleavage furrow
{ECO:0000269|PubMed:17956333}. Cell projection, lamellipodium
{ECO:0000269|PubMed:11919189}. Cell projection, ruffle
{ECO:0000269|PubMed:11919189}. Cell junction, focal adhesion
{ECO:0000269|PubMed:17158881}. Cell surface
{ECO:0000269|PubMed:17158881}. Note=Isoform 2 does not localize to
focal adhesions. Highly enriched in stage I melanosomes. Located
on plasma membrane of neuroblastoma NMB7 cells. In a lung cancer
cell line, in prometaphase and metaphase, localizes diffusely at
the membrane and in a few intracellular vesicles. In early
telophase, detected mainly on the matrix-facing side of the cells.
By mid-telophase, concentrated to the ingressing cleavage furrow,
mainly to the basal side of the furrow. In late telophase,
concentrated to the extending protrusions formed at the opposite
ends of the spreading daughter cells, in vesicles at the base of
the lamellipodia formed by the separating daughter cells.
Colocalizes with ITGB1BP1 and metastatic suppressor protein NME2
at the edge or peripheral ruffles and lamellipodia during the
early stages of cell spreading on fibronectin or collagen.
Translocates from peripheral focal adhesions sites to fibrillar
adhesions in a ITGB1BP1-dependent manner. Enriched preferentially
at invadopodia, cell membrane protrusions that correspond to sites
of cell invasion, in a collagen-dependent manner. Localized at
plasma and ruffle membranes in a collagen-independent manner.
{ECO:0000269|PubMed:10455171, ECO:0000303|PubMed:10455171}.
SUBCELLULAR LOCATION: Isoform 5: Cell membrane, sarcolemma
{ECO:0000250}. Cell junction {ECO:0000250}. Note=In cardiac
muscle, isoform 5 is found in costameres and intercalated disks.
{ECO:0000250}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CD320_HUMAN FUNCTION: Receptor for transcobalamin saturated with cobalamin
(TCbl) (PubMed:18779389). Plays an important role in cobalamin
uptake (PubMed:18779389, PubMed:20524213). Plasma membrane protein
that is expressed on follicular dendritic cells (FDC) and mediates
interaction with germinal center B cells (PubMed:10727470).
Functions as costimulator to promote B cell responses to antigenic
stimuli; promotes B cell differentiation and proliferation
(PubMed:10727470, PubMed:11418631). Germinal center-B (GC-B) cells
differentiate into memory B-cells and plasma cells (PC) through
interaction with T-cells and follicular dendritic cells (FDC)
(PubMed:11418631). CD320 augments the proliferation of PC
precursors generated by IL-10 (PubMed:11418631).
{ECO:0000269|PubMed:10727470, ECO:0000269|PubMed:11418631,
ECO:0000269|PubMed:18779389, ECO:0000269|PubMed:20524213}.
SUBCELLULAR LOCATION: Cell membrane {ECO:0000269|PubMed:10727470,
ECO:0000269|PubMed:11418631, ECO:0000305|PubMed:18779389}; Single-
pass type I membrane protein {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: PCD10_HUMAN FUNCTION: Potential calcium-dependent cell-adhesion protein.
SUBCELLULAR LOCATION: Cell membrane {ECO:0000250}; Single-pass
type I membrane protein {ECO:0000250}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CALR_HUMAN FUNCTION: Calcium-binding chaperone that promotes folding,
oligomeric assembly and quality control in the endoplasmic
reticulum (ER) via the calreticulin/calnexin cycle. This lectin
interacts transiently with almost all of the monoglucosylated
glycoproteins that are synthesized in the ER. Interacts with the
DNA-binding domain of NR3C1 and mediates its nuclear export.
Involved in maternal gene expression regulation. May participate
in oocyte maturation via the regulation of calcium homeostasis (By
similarity). {ECO:0000250, ECO:0000269|PubMed:11149926,
ECO:0000269|PubMed:7876246}.
SUBCELLULAR LOCATION: Endoplasmic reticulum lumen
{ECO:0000269|PubMed:10358038, ECO:0000269|PubMed:11149926}.
Cytoplasm, cytosol {ECO:0000269|PubMed:11149926}. Secreted,
extracellular space, extracellular matrix {ECO:0000305}. Cell
surface {ECO:0000269|PubMed:10358038}. Sarcoplasmic reticulum
lumen {ECO:0000250|UniProtKB:P28491}. Note=Also found in cell
surface (T cells), cytosol and extracellular matrix
(PubMed:10358038). Associated with the lytic granules in the
cytolytic T-lymphocytes. {ECO:0000269|PubMed:10358038,
ECO:0000269|PubMed:8418194}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: S39AE_HUMAN FUNCTION: Broad-scope metal ion transporter with a preference for
zinc uptake. Also mediates cellular uptake of nontransferrin-bound
iron. {ECO:0000269|PubMed:15642354, ECO:0000269|PubMed:27231142}.
SUBCELLULAR LOCATION: Cell membrane {ECO:0000269|PubMed:27231142};
Multi-pass membrane protein {ECO:0000255}. Cytoplasm
{ECO:0000269|PubMed:27231142}. Cell projection, lamellipodium
{ECO:0000269|PubMed:12659941, ECO:0000269|PubMed:15642354}.
Note=Localized to the plasma membrane and also found colocalized
with F-actin concentrated on lamellipodiae.
{ECO:0000269|PubMed:12659941, ECO:0000269|PubMed:15642354,
ECO:0000269|PubMed:27231142}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CLCC1_HUMAN FUNCTION: Seems to act as a chloride ion channel. {ECO:0000250}.
SUBCELLULAR LOCATION: Membrane {ECO:0000305}; Multi-pass membrane
protein {ECO:0000305}. Endoplasmic reticulum. Golgi apparatus.
Nucleus {ECO:0000250}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: C99L2_HUMAN FUNCTION: Plays a role in a late step of leukocyte extravasation
helping cells to overcome the endothelial basement membrane. Acts
at the same site as, but independently of, PECAM1 (By similarity).
Homophilic adhesion molecule, but these interactions may not be
required for cell aggregation (By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Cell membrane {ECO:0000250}; Single-pass
type I membrane protein {ECO:0000250}; Extracellular side
{ECO:0000250}. Cell junction {ECO:0000250}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: EMC1_HUMAN SUBCELLULAR LOCATION: Membrane {ECO:0000269|PubMed:22119785};
Single-pass type I membrane protein {ECO:0000269|PubMed:22119785}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: ROBO1_HUMAN FUNCTION: Receptor for SLIT1 and SLIT2 that mediates cellular
responses to molecular guidance cues in cellular migration,
including axonal navigation at the ventral midline of the neural
tube and projection of axons to different regions during neuronal
development (PubMed:10102268, PubMed:24560577). Interaction with
the intracellular domain of FLRT3 mediates axon attraction towards
cells expressing NTN1 (PubMed:24560577). In axon growth cones, the
silencing of the attractive effect of NTN1 by SLIT2 may require
the formation of a ROBO1-D complex (By similarity). Plays a role
in the regulation of cell migration via its interaction with
MYO9B; inhibits MYO9B-mediated stimulation of RHOA GTPase
activity, and thereby leads to increased levels of active, GTP-
bound RHOA (PubMed:26529257). May be required for lung development
(By similarity). {ECO:0000250|UniProtKB:O89026,
ECO:0000269|PubMed:10102268, ECO:0000269|PubMed:24560577,
ECO:0000269|PubMed:26529257, ECO:0000305}.
SUBCELLULAR LOCATION: Cell membrane {ECO:0000269|PubMed:24560577};
Single-pass type I membrane protein {ECO:0000305}. Cell
projection, axon {ECO:0000250|UniProtKB:O89026}. Note=Detected at
growth cones in thalamus neurons. {ECO:0000250|UniProtKB:O89026}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: PRDX4_HUMAN FUNCTION: Thiol-specific peroxidase that catalyzes the reduction
of hydrogen peroxide and organic hydroperoxides to water and
alcohols, respectively. Plays a role in cell protection against
oxidative stress by detoxifying peroxides and as sensor of
hydrogen peroxide-mediated signaling events. Regulates the
activation of NF-kappa-B in the cytosol by a modulation of I-
kappa-B-alpha phosphorylation. {ECO:0000269|PubMed:9388242}.
SUBCELLULAR LOCATION: Cytoplasm {ECO:0000269|PubMed:18052930,
ECO:0000269|PubMed:9388242}. Endoplasmic reticulum
{ECO:0000269|PubMed:18052930}. Note=Cotranslationally translocated
to and retained within the endoplasmic reticulum. A small fraction
of the protein is cytoplasmic. {ECO:0000269|PubMed:18052930}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: DAG1_HUMAN FUNCTION: The dystroglycan complex is involved in a number of
processes including laminin and basement membrane assembly,
sarcolemmal stability, cell survival, peripheral nerve
myelination, nodal structure, cell migration, and epithelial
polarization.
FUNCTION: Alpha-dystroglycan is an extracellular peripheral
glycoprotein that acts as a receptor for both extracellular matrix
proteins containing laminin-G domains. Receptor for laminin-2
(LAMA2) and agrin in peripheral nerve Schwann cells.
FUNCTION: Beta-dystroglycan is a transmembrane protein that plays
important roles in connecting the extracellular matrix to the
cytoskeleton. Acts as a cell adhesion receptor in both muscle and
non-muscle tissues. Receptor for both DMD and UTRN and, through
these interactions, scaffolds axin to the cytoskeleton. Also
functions in cell adhesion-mediated signaling and implicated in
cell polarity.
FUNCTION: (Microbial infection) Alpha-dystroglycan acts as a
receptor for lassa virus and lymphocytic choriomeningitis virus
glycoprotein and class C new-world arenaviruses (PubMed:16254364,
PubMed:19324387, PubMed:17360738). Alpha-dystroglycan acts as a
Schwann cell receptor for Mycobacterium leprae, the causative
organism of leprosy, but only in the presence of the G-domain of
LAMA2 (PubMed:9851927). {ECO:0000269|PubMed:16254364,
ECO:0000269|PubMed:17360738, ECO:0000269|PubMed:19324387,
ECO:0000269|PubMed:9851927}.
SUBCELLULAR LOCATION: Alpha-dystroglycan: Secreted, extracellular
space.
SUBCELLULAR LOCATION: Beta-dystroglycan: Cell membrane; Single-
pass type I membrane protein. Cytoplasm, cytoskeleton. Nucleus,
nucleoplasm. Cell membrane, sarcolemma {ECO:0000250}. Cell
junction, synapse, postsynaptic cell membrane {ECO:0000250}.
Note=The monomeric form translocates to the nucleus via the action
of importins and depends on RAN. Nuclear transport is inhibited by
Tyr-892 phosphorylation. In skeletal muscle, this phosphorylated
form locates to a vesicular internal membrane compartment. In
muscle cells, sarcolemma localization requires the presence of
ANK2, while localization to costameres requires the presence of
ANK3. Localizes to neuromuscular junctions (NMJs) in the presence
of ANK2 (By similarity). In peripheral nerves, localizes to the
Schwann cell membrane. Colocalizes with ERM proteins in Schwann-
cell microvilli. {ECO:0000250}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CNTN1_HUMAN FUNCTION: Contactins mediate cell surface interactions during
nervous system development. Involved in the formation of paranodal
axo-glial junctions in myelinated peripheral nerves and in the
signaling between axons and myelinating glial cells via its
association with CNTNAP1. Participates in oligodendrocytes
generation by acting as a ligand of NOTCH1. Its association with
NOTCH1 promotes NOTCH1 activation through the released notch
intracellular domain (NICD) and subsequent translocation to the
nucleus. Interaction with TNR induces a repulsion of neurons and
an inhibition of neurite outgrowth (By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Isoform 1: Cell membrane; Lipid-anchor, GPI-
anchor; Extracellular side.
SUBCELLULAR LOCATION: Isoform 2: Cell membrane; Lipid-anchor, GPI-
anchor; Extracellular side.
CLEAVAGE EVENTS OBSERVED:Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' RPN1_HUMAN SASSEAPPLINEDVK 23 PAPG | SASS RPN1_HUMAN SSEAPPLINEDVKR 25 PGSA | SSEA RPN1_HUMAN ASSEAPPLINEDVK 24 APGS | ASSE RPN1_HUMAN SEAPPLINEDVKR 26 GSAS | SEAP 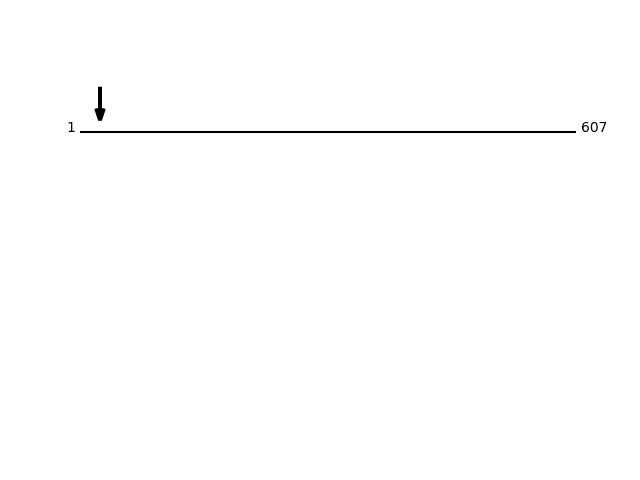
P68363 - Tubulin alpha-1B chainProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TBA1B_HUMAN SIQFVDWCPTGFK 340 KTKR | SIQF 
P19022 - Cadherin-2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CADH2_HUMAN LTTFTAQDPDR 516 AGTM | LTTF CADH2_HUMAN TFYGEVPENR 386 FTAM | TFYG CADH2_HUMAN AFDLPLSPVTIK 638 AGPF | AFDL CADH2_HUMAN TVAAENQVPLAK 461 MFVL | TVAA CADH2_HUMAN TTFTAQDPDR 517 GTML | TTFT CADH2_HUMAN YTKLSDPANWLK 534 QNIR | YTKL CADH2_HUMAN FYGEVPENR 387 TAMT | FYGE CADH2_HUMAN SLKPTLTEESVK 122 AVKL | SLKP CADH2_HUMAN AENQVPLAK 464 LTVA | AENQ CADH2_HUMAN AAENQVPLAK 463 VLTV | AAEN CADH2_HUMAN SDPANWLK 538 YTKL | SDPA CADH2_HUMAN FTAQDPDR 519 MLTT | FTAQ CADH2_HUMAN SGEIALCK 26 SVEA | SGEI CADH2_HUMAN YGEVPENR 388 AMTF | YGEV CADH2_HUMAN VTVIDVNENPYFAPNPK 486 ATVS | VTVI CADH2_HUMAN VIDVNENPYFAPNPK 488 VSVT | VIDV CADH2_HUMAN VAAENQVPLAK 462 FVLT | VAAE CADH2_HUMAN EVPIIITDSGNPPK 677 AGIY | EVPI CADH2_HUMAN GTMLTTFTAQDPDR 513 GLHA | GTML CADH2_HUMAN AVLDRESPNVK 557 ITTI | AVLD CADH2_HUMAN FAIQTDPNSNDGLVTVVKPIDFETNR 431 PTGR | FAIQ 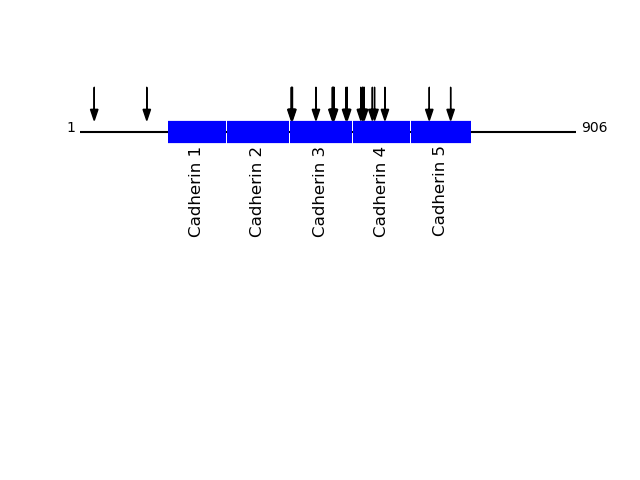
P11021 - 78 kDa glucose-regulated proteinProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' GRP78_HUMAN VYEGERPLTK 465 VTIK | VYEG GRP78_HUMAN TWNDPSVQQDIK 102 LIGR | TWND GRP78_HUMAN GIDLGTTYSCVGVFK 32 GTVV | GIDL GRP78_HUMAN YVAFTPEGER 65 ITPS | YVAF GRP78_HUMAN GSAGPPPTGEEDTAEKDEL 636 SKLY | GSAG GRP78_HUMAN AYGLDKR 208 AAAI | AYGL GRP78_HUMAN SCVGVFK 40 GTTY | SCVG 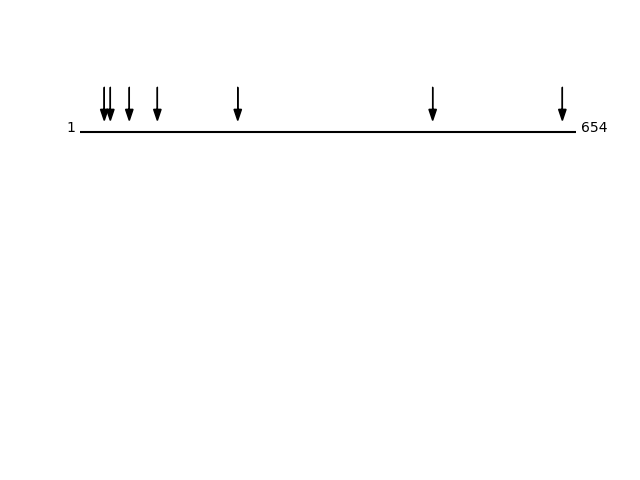
P27824 - CalnexinProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CALX_HUMAN WEVEEMK 104 YDGK | WEVE CALX_HUMAN HDGHDDDVIDIEDDLDDVIEEVEDSKPDTTAPPSSPK 21 IVEA | HDGH 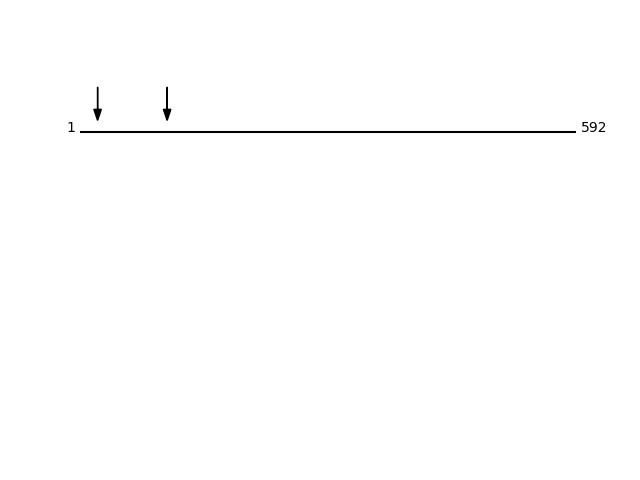
P14625 - EndoplasminProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' ENPL_HUMAN SLNIDPDAK 746 MLRL | SLNI ENPL_HUMAN EGVKFDESEK 594 NVAK | EGVK ENPL_HUMAN GWSGNMER 653 ASQY | GWSG ENPL_HUMAN NIDPDAK 748 RLSL | NIDP ENPL_HUMAN EFEPLLNWMK 614 AVEK | EFEP ENPL_HUMAN ALPEFDGK 579 YCIQ | ALPE ENPL_HUMAN AYQTGKDISTNYYASQKK 666 MKAQ | AYQT 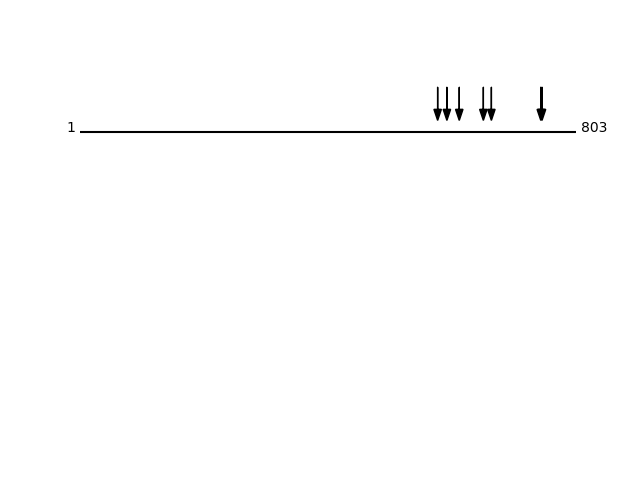
P01892 - HLA class I histocompatibility antigen, A-2 alpha chainProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' 1A02_HUMAN SFYPAEITLTWQR 231 CWAL | SFYP 1A02_HUMAN GYVDDTQFVR 50 FIAV | GYVD 1A02_HUMAN AAVVVPSGQEQR 269 FQKW | AAVV 1A02_HUMAN TAADMAAQTTK 158 LRSW | TAAD 1A02_HUMAN LEGTCVEWLR 184 LRAY | LEGT 1A02_HUMAN WAAVVVPSGQEQR 268 TFQK | WAAV 1A02_HUMAN AVVVPSGQEQR 270 QKWA | AVVV 1A02_HUMAN VVVPSGQEQR 271 KWAA | VVVP 1A02_HUMAN VVPSGQEQR 272 WAAV | VVPS 1A02_HUMAN SWTAADMAAQTTK 156 EDLR | SWTA 1A02_HUMAN FDSDAASQR 60 QFVR | FDSD 1A02_HUMAN YVDDTQFVR 51 IAVG | YVDD 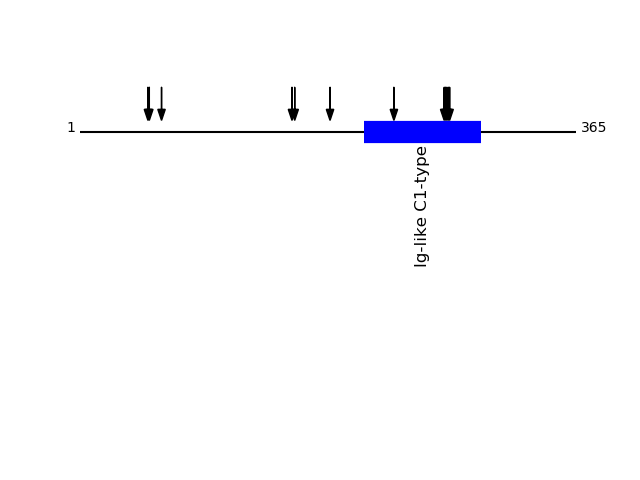
P14209 - CD99 antigenProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CD99_HUMAN GSFSDADLADGVSGGEGK 88 PSSS | GSFS CD99_HUMAN NHPSSSGSFSDADLADGVSGGEGK 82 NPNP | NHPS CD99_HUMAN FSDADLADGVSGGEGK 90 SSGS | FSDA CD99_HUMAN SFSDADLADGVSGGEGK 89 SSSG | SFSD CD99_HUMAN SSGSFSDADLADGVSGGEGK 86 NHPS | SSGS CD99_HUMAN SGSFSDADLADGVSGGEGK 87 HPSS | SGSF CD99_HUMAN SDADLADGVSGGEGK 91 SGSF | SDAD CD99_HUMAN DADLADGVSGGEGK 92 GSFS | DADL CD99_HUMAN LADGVSGGEGK 95 SDAD | LADG CD99_HUMAN SSSGSFSDADLADGVSGGEGK 85 PNHP | SSSG 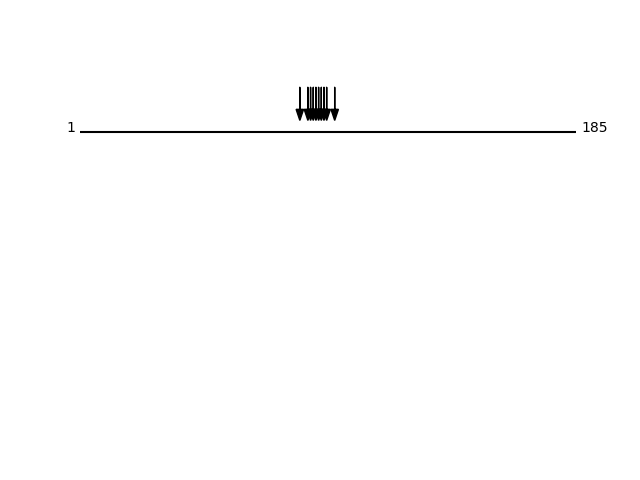
P10586 - Receptor-type tyrosine-protein phosphatase FProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' PTPRF_HUMAN AHTDVGPGPESSPVLVR 686 VWVR | AHTD PTPRF_HUMAN SVAYEAVDGEDR 643 ITQY | SVAY PTPRF_HUMAN AFTAVGDGPPSPTIQVK 490 LRVL | AFTA PTPRF_HUMAN GDGPPSPTIQVK 495 FTAV | GDGP PTPRF_HUMAN GSGPLSPSIQSR 995 WTSK | GSGP PTPRF_HUMAN AVGDGPPSPTIQVK 493 LAFT | AVGD PTPRF_HUMAN HTDVGPGPESSPVLVR 687 WVRA | HTDV 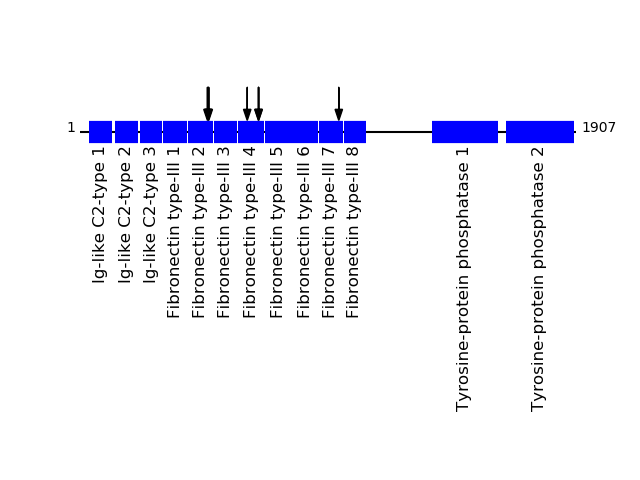
P68104 - Elongation factor 1-alpha 1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' EF1A1_HUMAN SFKYAWVLDK 53 MGKG | SFKY 
P35613 - BasiginProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' BASI_HUMAN LNMEADPGQYR 290 HIEN | LNME BASI_HUMAN SVPPVTDWAWYK 246 CKSE | SVPP BASI_HUMAN NIQLHGPPR 214 MGTA | NIQL BASI_HUMAN NMEADPGQYR 291 IENL | NMEA BASI_HUMAN LHIENLNMEADPGQYR 285 GRSE | LHIE BASI_HUMAN NLNMEADPGQYR 289 LHIE | NLNM BASI_HUMAN IQLHGPPR 215 GTAN | IQLH 
P06576 - ATP synthase subunit beta, mitochondrialProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' ATPB_HUMAN AAQTSPSPK 47 VRDY | AAQT 
Q9Y4L1 - Hypoxia up-regulated protein 1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' HYOU1_HUMAN GNTISSLFGGGTTPDAK 578 LTKL | GNTI HYOU1_HUMAN GVAPAPEGEK 676 AGPE | GVAP HYOU1_HUMAN VVLDLPDLPEDK 704 GVEL | VVLD HYOU1_HUMAN VMSVDLGSESMK 35 DTLA | VMSV HYOU1_HUMAN GLMDDVDFK 332 AQIE | GLMD HYOU1_HUMAN SSLFGGGTTPDAK 582 GNTI | SSLF HYOU1_HUMAN SYGVFR 212 ATAL | SYGV HYOU1_HUMAN SLFGGGTTPDAK 583 NTIS | SLFG HYOU1_HUMAN LAVMSVDLGSESMK 33 LSDT | LAVM 
Q15758 - Neutral amino acid transporter B(0)Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' AAAT_HUMAN SIEDQGAAAGGYCGSR 27 LALA | SIED AAAT_HUMAN SVGAAGSAENAPSK 165 AINA | SVGA AAAT_HUMAN MVADPPR 1 - | MVAD AAAT_HUMAN GSAENAPSKEVLDSFLDLAR 170 VGAA | GSAE 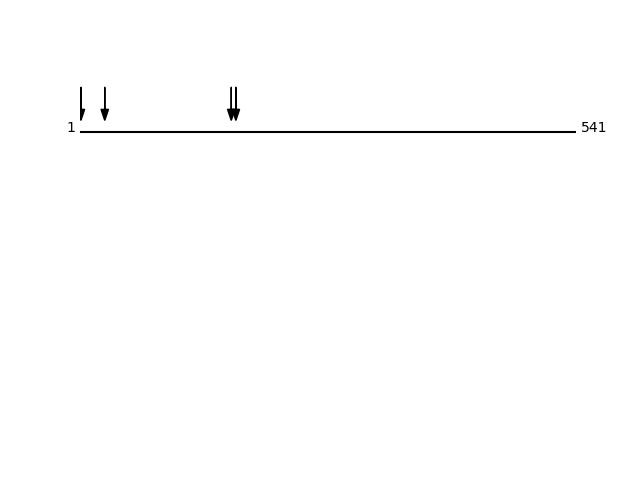
Q14126 - Desmoglein-2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' DSG2_HUMAN GNFQAFDEDTGLPAHAR 411 GQII | GNFQ DSG2_HUMAN AFDEDTGLPAHAR 415 GNFQ | AFDE DSG2_HUMAN SLEPAYPPVFYLNK 202 YRIV | SLEP DSG2_HUMAN VCECLHGSGCR 590 LTLT | VCEC DSG2_HUMAN AWITAPVALR 50 RQKR | AWIT DSG2_HUMAN LAKLPDFESR 449 SEIK | LAKL DSG2_HUMAN SKYKPTPIPIK 366 KSIR | SKYK DSG2_HUMAN TVCECLHGSGCR 589 VLTL | TVCE DSG2_HUMAN LHLQVLSTR 24 VGSG | LHLQ DSG2_HUMAN LISDNQGFSCPEK 571 EIQF | LISD DSG2_HUMAN AISEDYPR 471 VKIV | AISE DSG2_HUMAN ITAPVALR 52 KRAW | ITAP DSG2_HUMAN IVSLEPAYPPVFYLNK 200 ISYR | IVSL 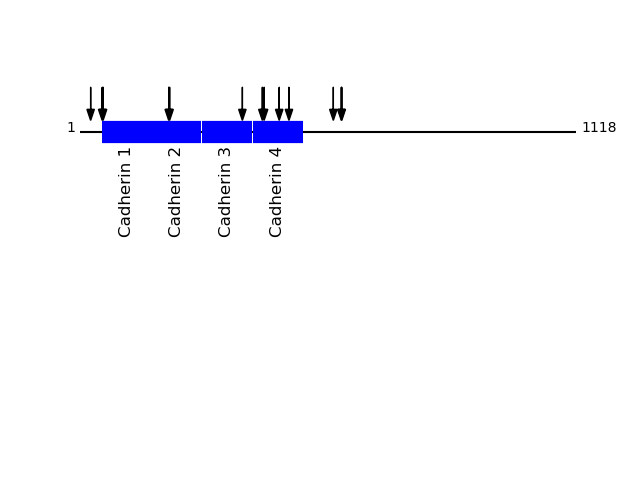
Q99805 - Transmembrane 9 superfamily member 2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TM9S2_HUMAN SVSFEEDDKIR 272 AYTY | SVSF TM9S2_HUMAN FYLPGLAPVNFCDEEKK 36 RSGA | FYLP TM9S2_HUMAN SFEEDDKIR 274 TYSV | SFEE 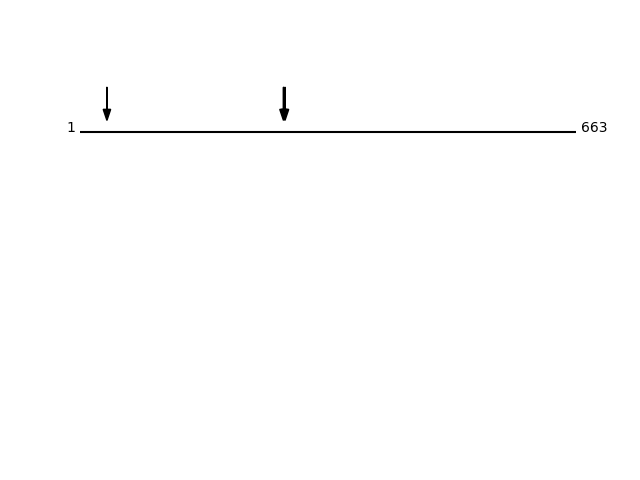
Q8NBS9 - Thioredoxin domain-containing protein 5Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TXND5_HUMAN AADGPPAADGEDGQDPHSK 45 AAAA | AADG TXND5_HUMAN AAADGPPAADGEDGQDPHSK 44 EAAA | AAAD 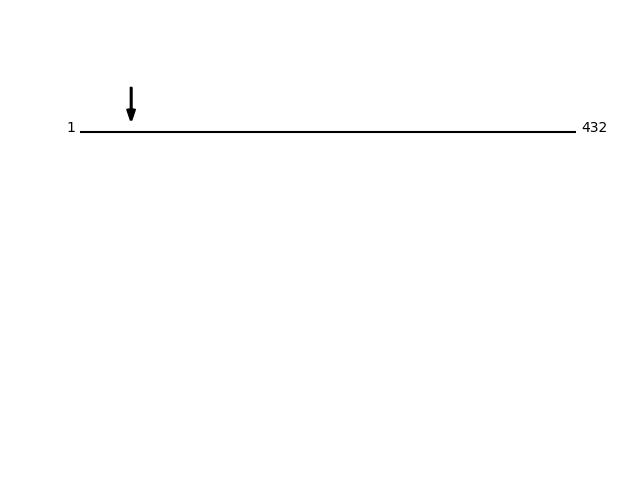
P16615 - Sarcoplasmic/endoplasmic reticulum calcium ATPase 2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' AT2A2_HUMAN GAPEGVIDR 515 MFVK | GAPE 
P04844 - Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' RPN2_HUMAN LHNQKTGQEVVFVAEPDNK 438 TFVR | LHNQ 
Q13740 - CD166 antigenProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CD166_HUMAN SIPEHDEADEISDENR 504 VSAI | SIPE CD166_HUMAN AISIPEHDEADEISDENR 502 LNVS | AISI CD166_HUMAN LVTEDNVFEAPTIVK 115 FVCM | LVTE CD166_HUMAN ALFLETEQLK 143 IVSK | ALFL CD166_HUMAN SVTYYGPSGQK 221 PFTC | SVTY CD166_HUMAN YYGPSGQK 224 CSVT | YYGP CD166_HUMAN HYQDAGNYVCETALQEVEGLK 383 FSSL | HYQD 
P14314 - Glucosidase 2 subunit betaProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' GLU2B_HUMAN ACPEPPPEAPTEDDHDEL 511 MTPA | ACPE GLU2B_HUMAN ALLSGDTQTDATSFYDR 255 AEAQ | ALLS GLU2B_HUMAN AQQEQELAADAFK 207 AAAK | AQQE GLU2B_HUMAN ALPTDLPAPSAPDLTEPK 284 YRSE | ALPT GLU2B_HUMAN LSGDTQTDATSFYDR 257 AQAL | LSGD GLU2B_HUMAN LLSGDTQTDATSFYDR 256 EAQA | LLSG GLU2B_HUMAN SLEDQVEMLR 168 AGKK | SLED 
Q5JRA6 - Transport and Golgi organization protein 1 homolog {ECO:0000305}Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TGO1_HUMAN MAPPLEEGLGGAMEEMQPLHEDNFSR 1043 ATLV | MAPP TGO1_HUMAN GLAGEPEGELSKEDHENTEK 866 LKTS | GLAG 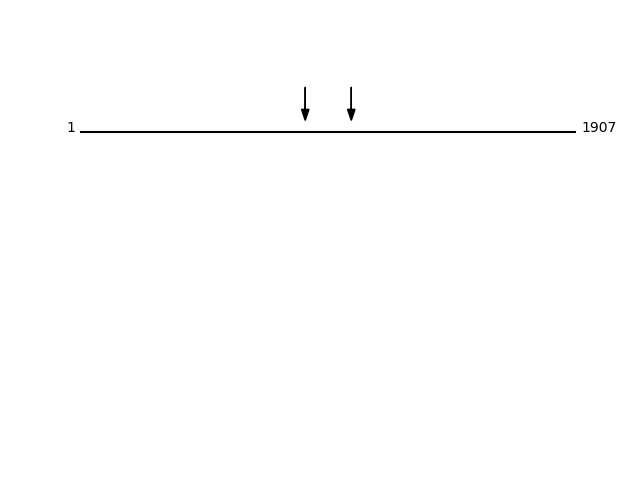
O14763 - Tumor necrosis factor receptor superfamily member 10BProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TR10B_HUMAN ALITQQDLAPQQR 54 SAES | ALIT TR10B_HUMAN LITQQDLAPQQR 55 AESA | LITQ TR10B_HUMAN ITQQDLAPQQR 56 ESAL | ITQQ TR10B_HUMAN QQDLAPQQR 58 ALIT | QQDL 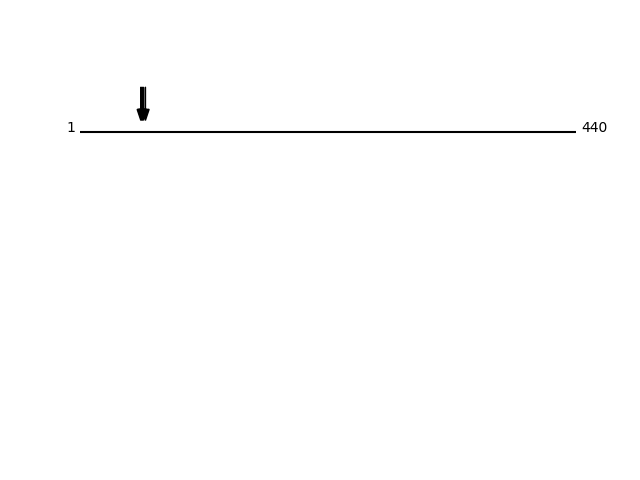
P08195 - 4F2 cell-surface antigen heavy chainProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' 4F2_HUMAN GQSEDPGSLLSLFR 511 MTVK | GQSE 4F2_HUMAN SYLSDSGSTGEHTK 403 LLTS | SYLS 4F2_HUMAN VAEDEAEAAAAAK 148 VKIK | VAED 4F2_HUMAN GLVLGPIHK 258 LKVK | GLVL 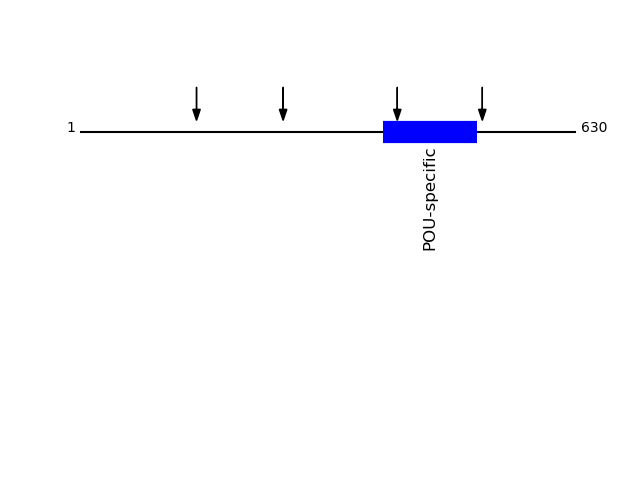
P05556 - Integrin beta-1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' ITB1_HUMAN FCECDNFNCDR 552 YSGK | FCEC ITB1_HUMAN GCPPDDIENPR 74 LKKK | GCPP 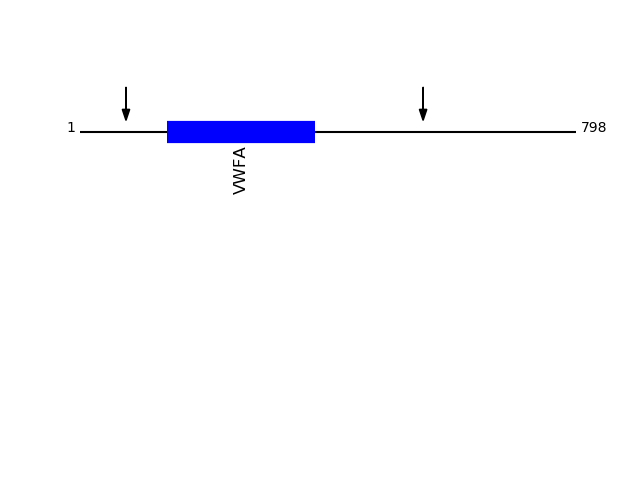
Q9NPF0 - CD320 antigenProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CD320_HUMAN AASPLSTPTSAQAAGPSSGSCPPTK 34 GLEA | AASP CD320_HUMAN AAGPSSGSCPPTK 46 TSAQ | AAGP CD320_HUMAN STPTSAQAAGPSSGSCPPTK 39 ASPL | STPT 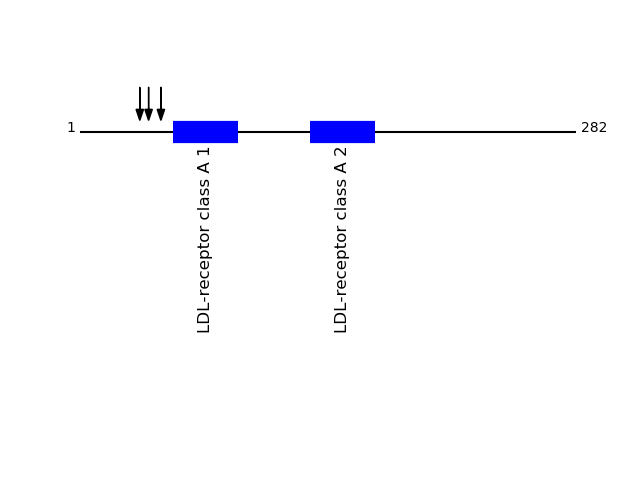
Q9P2E7 - Protocadherin-10Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' PCD10_HUMAN LVDGAVEPQGGGGSGGGGSGEHQRPSR 678 LVVQ | LVDG PCD10_HUMAN VAAVDADDGENAR 603 LLTR | VAAV PCD10_HUMAN AAVDADDGENAR 604 LTRV | AAVD PCD10_HUMAN AVDADDGENAR 605 TRVA | AVDA PCD10_HUMAN SIQVQVSDVNDNAPR 448 STSK | SIQV 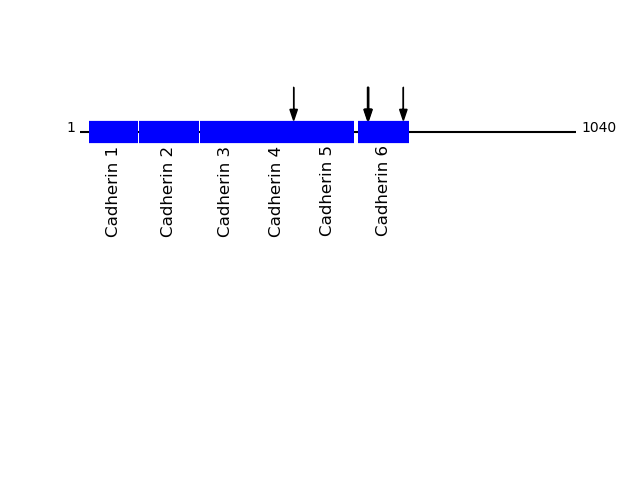
P27797 - CalreticulinProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CALR_HUMAN SASFEPFSNK 78 FYAL | SASF CALR_HUMAN SFEPFSNK 80 ALSA | SFEP 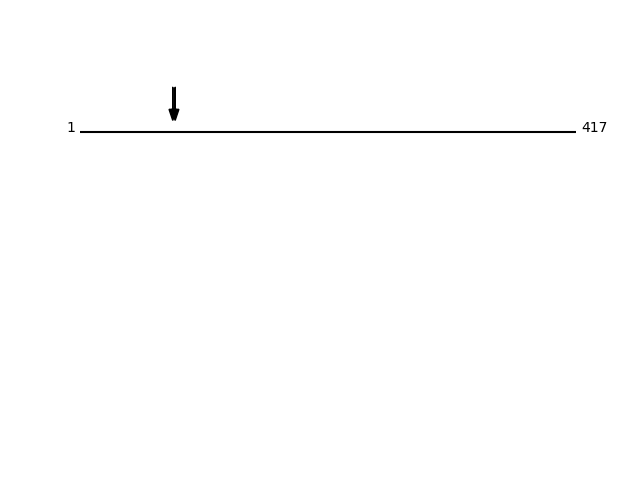
Q15043 - Zinc transporter ZIP14Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' S39AE_HUMAN SSLGAPAISAASFLQDLIHR 31 EAHA | SSLG S39AE_HUMAN GAPAISAASFLQDLIHR 34 ASSL | GAPA 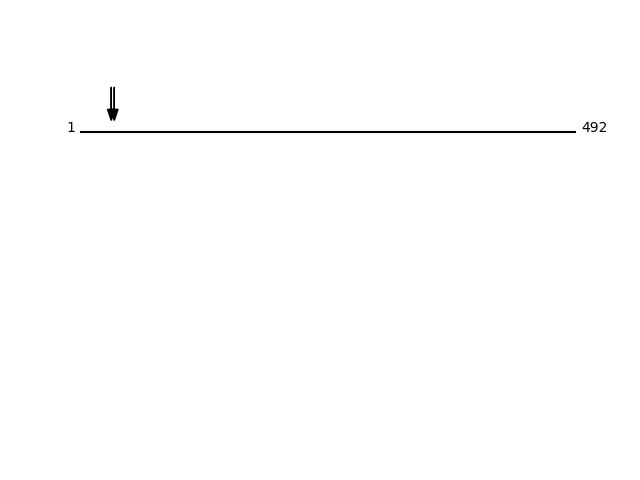
Q96S66 - Chloride channel CLIC-like protein 1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CLCC1_HUMAN YAHDDDWIDPTDMLNYDAASGTMR 17 LVAG | YAHD CLCC1_HUMAN HDDDWIDPTDMLNYDAASGTMR 19 AGYA | HDDD CLCC1_HUMAN GISGEKDVSPDLSCADEISECYHK 47 QAKY | GISG 
Q8TCZ2 - CD99 antigen-like protein 2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' C99L2_HUMAN GNDFDLADALDDRNDR 116 ANTL | GNDF C99L2_HUMAN DFDDFNLEDAVK 26 RGSG | DFDD C99L2_HUMAN GLDLADALDDQDDGR 70 PPGS | GLDL C99L2_HUMAN GSGLDLADALDDQDDGR 68 AKPP | GSGL C99L2_HUMAN ALDDQDDGR 76 DLAD | ALDD C99L2_HUMAN GGGGFSDKDLEDIVGGGEYKPDK 141 KPIA | GGGG C99L2_HUMAN GFSDKDLEDIVGGGEYKPDK 144 AGGG | GFSD 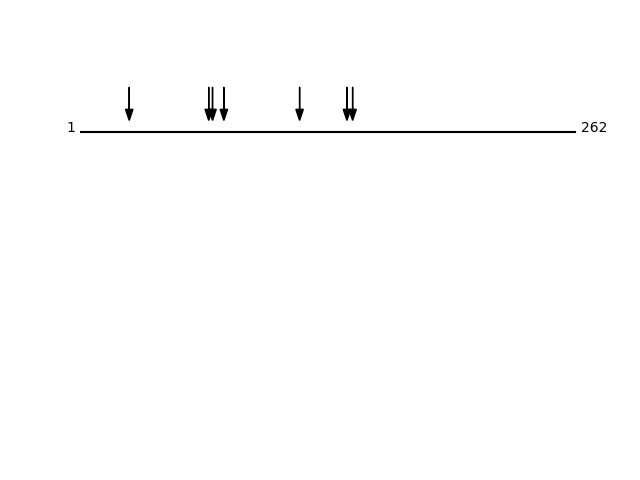
Q8N766 - ER membrane protein complex subunit 1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' EMC1_HUMAN SSSEDGSMGSFSEK 346 VQKS | SSSE EMC1_HUMAN KDDSVGYR 413 VFLK | KDDS EMC1_HUMAN VYEDQVGK 22 PAAA | VYED 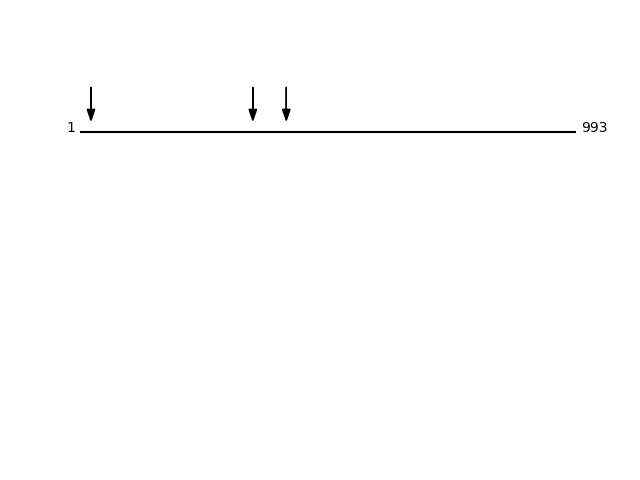
Q9Y6N7 - Roundabout homolog 1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' ROBO1_HUMAN FNEFQGADSEIK 755 ARPF | FNEF ROBO1_HUMAN GEPAVMECQPPR 184 MVAV | GEPA ROBO1_HUMAN FQGADSEIK 758 FFNE | FQGA ROBO1_HUMAN GTNMVGER 242 YVCV | GTNM ROBO1_HUMAN AYLEVTDVIADRPPPVIR 441 IITK | AYLE ROBO1_HUMAN VCVGTNMVGER 239 AGKY | VCVG ROBO1_HUMAN ARPFFNEFQGADSEIK 751 YEIK | ARPF ROBO1_HUMAN EFQGADSEIK 757 PFFN | EFQG 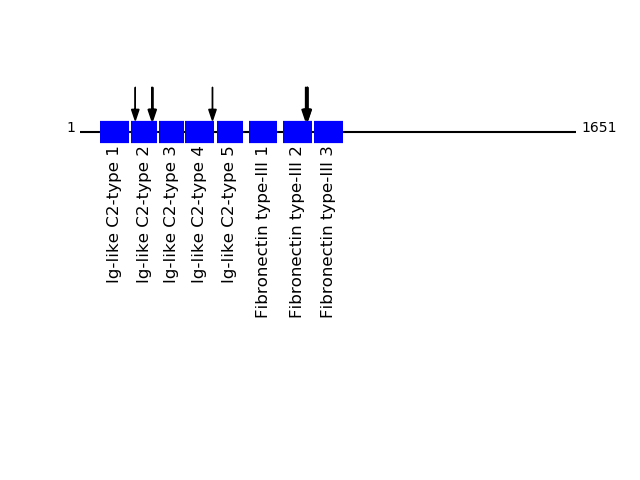
Q13162 - Peroxiredoxin-4Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' PRDX4_HUMAN VQGWETEERPR 35 PAGA | VQGW PRDX4_HUMAN WETEERPR 38 AVQG | WETE PRDX4_HUMAN GWETEERPR 37 GAVQ | GWET PRDX4_HUMAN GQVYPGEASR 57 FYAG | GQVY 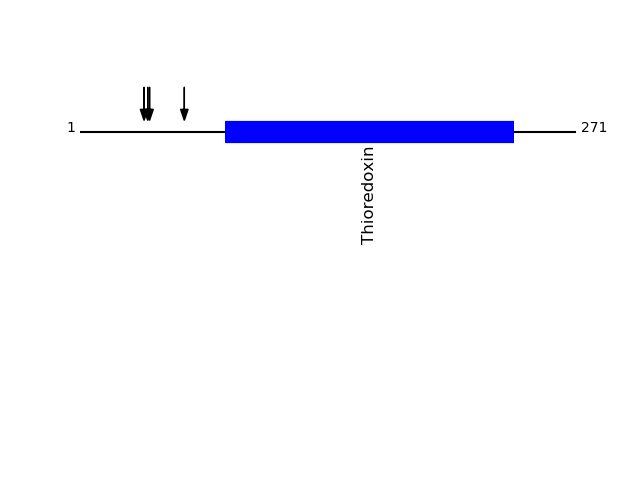
Q14118 - DystroglycanProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' DAG1_HUMAN FIPVVPPR 718 RHLQ | FIPV DAG1_HUMAN LQFIPVVPPR 716 SCRH | LQFI DAG1_HUMAN QFIPVVPPR 717 CRHL | QFIP DAG1_HUMAN HWPSEPSEAVR 30 MAQS | HWPS 
Q12860 - Contactin-1