Listed below are N-terminal peptides that were captured and enriched from the surface of a HEK293T cell line expressing subtiligase-Y217D-TM.
RPN1_HUMAN
P04843 - Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1
FUNCTION: Essential subunit of the N-oligosaccharyl transferase (OST) complex which catalyzes the transfer of a high mannose oligosaccharide from a lipid-linked oligosaccharide donor to an asparagine residue within an Asn-X-Ser/Thr consensus motif in nascent polypeptide chains. {ECO:0000250|UniProtKB:E2RQ08, ECO:0000250|UniProtKB:Q9GMB0, ECO:0000305}.
SUBCELLULAR LOCATION: Endoplasmic reticulum {ECO:0000250|UniProtKB:E2RQ08, ECO:0000250|UniProtKB:Q9GMB0}. Endoplasmic reticulum membrane; Single-pass type I membrane protein {ECO:0000305}. Melanosome. Note=Identified by mass spectrometry in melanosome fractions from stage I to stage IV.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CADH2_HUMAN FUNCTION: Cadherins are calcium-dependent cell adhesion proteins.
They preferentially interact with themselves in a homophilic
manner in connecting cells; cadherins may thus contribute to the
sorting of heterogeneous cell types. Acts as a regulator of neural
stem cells quiescence by mediating anchorage of neural stem cells
to ependymocytes in the adult subependymal zone: upon cleavage by
MMP24, CDH2-mediated anchorage is affected, leading to modulate
neural stem cell quiescence. CDH2 may be involved in neuronal
recognition mechanism. In hippocampal neurons, may regulate
dendritic spine density (By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Cell membrane
{ECO:0000250|UniProtKB:P15116}; Single-pass type I membrane
protein {ECO:0000255}. Cell membrane, sarcolemma
{ECO:0000250|UniProtKB:P15116}. Cell junction
{ECO:0000269|PubMed:28169360}. Cell surface
{ECO:0000250|UniProtKB:P15116}. Note=Colocalizes with TMEM65 at
the intercalated disk in cardiomyocytes. Colocalizes with OBSCN at
the intercalated disk and at sarcolemma in cardiomyocytes.
{ECO:0000250|UniProtKB:P15116}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TXND5_HUMAN FUNCTION: Possesses thioredoxin activity. Has been shown to reduce
insulin disulfide bonds. Also complements protein disulfide-
isomerase deficiency in yeast (By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Endoplasmic reticulum lumen
{ECO:0000255|PROSITE-ProRule:PRU10138}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TM9S2_HUMAN FUNCTION: In the intracellular compartments, may function as a
channel or small molecule transporter.
SUBCELLULAR LOCATION: Endosome membrane {ECO:0000305}; Multi-pass
membrane protein {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CNPY4_HUMAN FUNCTION: Plays a role in the regulation of the cell surface
expression of TLR4. {ECO:0000269|PubMed:16338228}.
SUBCELLULAR LOCATION: Secreted {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: S39AA_HUMAN FUNCTION: May act as a zinc-influx transporter.
{ECO:0000269|PubMed:17359283}.
SUBCELLULAR LOCATION: Membrane {ECO:0000305}; Multi-pass membrane
protein {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TMX4_HUMAN SUBCELLULAR LOCATION: Membrane {ECO:0000305}; Single-pass type I
membrane protein {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: S39A7_HUMAN FUNCTION: Zinc transporter, that transports Zn(2+) from the
endoplasmic reticulum/Golgi apparatus to the cytosol. Transport is
stimulated by growth factors, such as EGF, and Ca(2+), as well as
by exogenous Zn(2+). {ECO:0000269|PubMed:14525538,
ECO:0000269|PubMed:15705588, ECO:0000269|PubMed:22317921}.
SUBCELLULAR LOCATION: Endoplasmic reticulum membrane; Multi-pass
membrane protein. Golgi apparatus, cis-Golgi network membrane.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: GLU2B_HUMAN FUNCTION: Regulatory subunit of glucosidase II.
{ECO:0000269|PubMed:10929008}.
SUBCELLULAR LOCATION: Endoplasmic reticulum {ECO:0000255|PROSITE-
ProRule:PRU10138}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: GPI8_HUMAN FUNCTION: Mediates GPI anchoring in the endoplasmic reticulum, by
replacing a protein's C-terminal GPI attachment signal peptide
with a pre-assembled GPI. During this transamidation reaction, the
GPI transamidase forms a carbonyl intermediate with the substrate
protein.
SUBCELLULAR LOCATION: Endoplasmic reticulum membrane
{ECO:0000269|PubMed:11483512}; Single-pass type I membrane protein
{ECO:0000269|PubMed:11483512}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: LRIG2_HUMAN SUBCELLULAR LOCATION: Cell membrane {ECO:0000269|PubMed:15145052};
Single-pass type I membrane protein {ECO:0000269|PubMed:15145052}.
Cytoplasm {ECO:0000269|PubMed:15145052}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TR10B_HUMAN FUNCTION: Receptor for the cytotoxic ligand TNFSF10/TRAIL
(PubMed:10549288). The adapter molecule FADD recruits caspase-8 to
the activated receptor. The resulting death-inducing signaling
complex (DISC) performs caspase-8 proteolytic activation which
initiates the subsequent cascade of caspases (aspartate-specific
cysteine proteases) mediating apoptosis. Promotes the activation
of NF-kappa-B. Essential for ER stress-induced apoptosis.
{ECO:0000269|PubMed:10542098, ECO:0000269|PubMed:10549288,
ECO:0000269|PubMed:15322075}.
SUBCELLULAR LOCATION: Membrane; Single-pass type I membrane
protein.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: SELT_HUMAN FUNCTION: Selenoprotein with thioredoxin reductase-like
oxidoreductase activity (By similarity). Protects dopaminergic
neurons against oxidative stress ans cell death (PubMed:26866473).
Involved in ADCYAP1/PACAP-induced calcium mobilization and
neuroendocrine secretion (By similarity). Plays a role in
fibroblast anchorage and redox regulation (By similarity). In
gastric smooth muscle, modulates the contraction processes through
the regulation of calcium release and MYLK activation (By
similarity). In pancreatic islets, involved in the control of
glucose homeostasis, contributes to prolonged ADCYAP1/PACAP-
induced insulin secretion (By similarity).
{ECO:0000250|UniProtKB:P62342, ECO:0000250|UniProtKB:Q1H5H1,
ECO:0000269|PubMed:26866473}.
SUBCELLULAR LOCATION: Endoplasmic reticulum membrane
{ECO:0000250|UniProtKB:Q1H5H1}; Single-pass membrane protein
{ECO:0000255}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: NOTC2_HUMAN FUNCTION: Functions as a receptor for membrane-bound ligands
Jagged1, Jagged2 and Delta1 to regulate cell-fate determination.
Upon ligand activation through the released notch intracellular
domain (NICD) it forms a transcriptional activator complex with
RBPJ/RBPSUH and activates genes of the enhancer of split locus.
Affects the implementation of differentiation, proliferation and
apoptotic programs (By similarity). Involved in bone remodeling
and homeostasis. In collaboration with RELA/p65 enhances NFATc1
promoter activity and positively regulates RANKL-induced
osteoclast differentiation. Positively regulates self-renewal of
liver cancer cells (PubMed:25985737).
{ECO:0000250|UniProtKB:O35516, ECO:0000269|PubMed:21378985,
ECO:0000269|PubMed:21378989, ECO:0000269|PubMed:25985737}.
SUBCELLULAR LOCATION: Notch 2 extracellular truncation: Cell
membrane {ECO:0000269|PubMed:9244302}; Single-pass type I membrane
protein {ECO:0000269|PubMed:9244302}.
SUBCELLULAR LOCATION: Notch 2 intracellular domain: Nucleus
{ECO:0000269|PubMed:25985737}. Cytoplasm
{ECO:0000269|PubMed:25985737}. Note=Following proteolytical
processing NICD is translocated to the nucleus. Retained at the
cytoplasm by C8orf4 (PubMed:25985737).
{ECO:0000269|PubMed:25985737}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: JAM1_HUMAN FUNCTION: Seems to play a role in epithelial tight junction
formation. Appears early in primordial forms of cell junctions and
recruits PARD3 (PubMed:11489913). The association of the PARD6-
PARD3 complex may prevent the interaction of PARD3 with JAM1,
thereby preventing tight junction assembly (By similarity). Plays
a role in regulating monocyte transmigration involved in integrity
of epithelial barrier (By similarity). Ligand for integrin alpha-
L/beta-2 involved in memory T-cell and neutrophil transmigration
(PubMed:11812992). Involved in platelet activation
(PubMed:10753840). {ECO:0000250|UniProtKB:O88792,
ECO:0000269|PubMed:10753840, ECO:0000269|PubMed:11489913,
ECO:0000269|PubMed:11812992}.
FUNCTION: (Microbial infection) Acts as a receptor for Mammalian
reovirus sigma-1. {ECO:0000269|PubMed:11239401}.
FUNCTION: (Microbial infection) Acts as a receptor for Human
Rotavirus strain Wa. {ECO:0000269|PubMed:25481868}.
SUBCELLULAR LOCATION: Cell junction, tight junction
{ECO:0000269|PubMed:11171323}. Cell membrane
{ECO:0000269|PubMed:11171323}; Single-pass type I membrane protein
{ECO:0000269|PubMed:11171323}. Note=Localized at tight junctions
of both epithelial and endothelial cells.
{ECO:0000269|PubMed:11171323}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: EMC7_HUMAN SUBCELLULAR LOCATION: Membrane {ECO:0000269|PubMed:22119785};
Single-pass type I membrane protein {ECO:0000269|PubMed:22119785}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TGO1_HUMAN FUNCTION: Plays a role in the transport of cargos that are too
large to fit into COPII-coated vesicles and require specific
mechanisms to be incorporated into membrane-bound carriers and
exported from the endoplasmic reticulum. This protein is required
for collagen VII (COL7A1) secretion by loading COL7A1 into
transport carriers. It may participate in cargo loading of COL7A1
at endoplasmic reticulum exit sites by binding to COPII coat
subunits Sec23/24 and guiding SH3-bound COL7A1 into a growing
carrier. Does not play a role in global protein secretion and is
apparently specific to COL7A1 cargo loading. However, it may
participate in secretion of other proteins in cells that do not
secrete COL7A1. It is also specifically required for the secretion
of lipoproteins by participating in their export from the
endoplasmic reticulum (PubMed:27138255).
{ECO:0000269|PubMed:19269366, ECO:0000269|PubMed:27138255}.
SUBCELLULAR LOCATION: Endoplasmic reticulum membrane
{ECO:0000269|PubMed:19269366, ECO:0000269|PubMed:21525241};
Single-pass membrane protein {ECO:0000269|PubMed:19269366}.
Note=Localizes at endoplasmic reticulum exit sites. After loading
of COL7A1 into transport carriers, it is not incorporated into
COPII carriers and remains in the endoplasmic reticulum membrane.
{ECO:0000269|PubMed:19269366, ECO:0000269|PubMed:21525241}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: SDC1_HUMAN FUNCTION: Cell surface proteoglycan that bears both heparan
sulfate and chondroitin sulfate and that links the cytoskeleton to
the interstitial matrix. Regulates exosome biogenesis in concert
with SDCBP and PDCD6IP (PubMed:22660413).
{ECO:0000269|PubMed:22660413}.
SUBCELLULAR LOCATION: Membrane {ECO:0000255}; Single-pass type I
membrane protein {ECO:0000255}. Secreted
{ECO:0000269|PubMed:9169435}. Secreted, exosome
{ECO:0000269|PubMed:22660413}. Note=Shedding of the ectodomain
produces a soluble form. {ECO:0000269|PubMed:9169435}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: STIM1_HUMAN FUNCTION: Plays a role in mediating store-operated Ca(2+) entry
(SOCE), a Ca(2+) influx following depletion of intracellular
Ca(2+) stores (PubMed:15866891, PubMed:16005298, PubMed:16208375,
PubMed:16537481, PubMed:16733527, PubMed:16766533,
PubMed:16807233, PubMed:18854159, PubMed:19249086,
PubMed:22464749, PubMed:24069340, PubMed:24351972,
PubMed:24591628, PubMed:26322679, PubMed:25326555,
PubMed:28219928). Acts as Ca(2+) sensor in the endoplasmic
reticulum via its EF-hand domain. Upon Ca(2+) depletion,
translocates from the endoplasmic reticulum to the plasma membrane
where it activates the Ca(2+) release-activated Ca(2+) (CRAC)
channel subunit ORAI1 (PubMed:16208375, PubMed:16537481). Involved
in enamel formation (PubMed:24621671). Activated following
interaction with TMEM110/STIMATE, leading to promote STIM1
conformational switch (PubMed:26322679).
{ECO:0000269|PubMed:15866891, ECO:0000269|PubMed:16005298,
ECO:0000269|PubMed:16208375, ECO:0000269|PubMed:16537481,
ECO:0000269|PubMed:16733527, ECO:0000269|PubMed:16766533,
ECO:0000269|PubMed:16807233, ECO:0000269|PubMed:18854159,
ECO:0000269|PubMed:19249086, ECO:0000269|PubMed:22464749,
ECO:0000269|PubMed:24069340, ECO:0000269|PubMed:24351972,
ECO:0000269|PubMed:24591628, ECO:0000269|PubMed:24621671,
ECO:0000269|PubMed:25326555, ECO:0000269|PubMed:26322679,
ECO:0000269|PubMed:28219928}.
SUBCELLULAR LOCATION: Cell membrane; Single-pass type I membrane
protein {ECO:0000269|PubMed:11004585, ECO:0000269|PubMed:16005298,
ECO:0000269|PubMed:16208375, ECO:0000269|PubMed:18854159,
ECO:0000269|PubMed:19249086, ECO:0000269|PubMed:28219928}.
Endoplasmic reticulum membrane; Single-pass type I membrane
protein {ECO:0000269|PubMed:16005298, ECO:0000269|PubMed:16208375,
ECO:0000269|PubMed:18854159, ECO:0000269|PubMed:19249086,
ECO:0000269|PubMed:26322679}. Cytoplasm, cytoskeleton
{ECO:0000269|PubMed:19632184}. Sarcoplasmic reticulum
{ECO:0000269|PubMed:25326555}. Note=Translocates from the
endoplasmic reticulum to the cell membrane in response to a
depletion of intracellular calcium and is detected at punctae
corresponding to junctions between the endoplasmic reticulum and
the cell membrane (PubMed:19249086, PubMed:16005298,
PubMed:16208375, PubMed:18854159). Associated with the microtubule
network at the growing distal tip of microtubules
(PubMed:19632184). {ECO:0000269|PubMed:16005298,
ECO:0000269|PubMed:16208375, ECO:0000269|PubMed:18854159,
ECO:0000269|PubMed:19249086, ECO:0000269|PubMed:19632184}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: OCLN_HUMAN FUNCTION: May play a role in the formation and regulation of the
tight junction (TJ) paracellular permeability barrier. It is able
to induce adhesion when expressed in cells lacking tight
junctions. {ECO:0000269|PubMed:19114660}.
SUBCELLULAR LOCATION: Cell membrane {ECO:0000269|PubMed:19017651,
ECO:0000269|PubMed:19114660, ECO:0000269|PubMed:9175707}; Multi-
pass membrane protein {ECO:0000255}. Cell junction, tight junction
{ECO:0000269|PubMed:10523508, ECO:0000269|PubMed:19017651,
ECO:0000269|PubMed:19114660, ECO:0000269|PubMed:19332538,
ECO:0000269|PubMed:9175707}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CD99_HUMAN FUNCTION: Involved in T-cell adhesion processes and in spontaneous
rosette formation with erythrocytes. Plays a role in a late step
of leukocyte extravasation helping leukocytes to overcome the
endothelial basement membrane. Acts at the same site as, but
independently of, PECAM1. Involved in T-cell adhesion processes
(By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Membrane {ECO:0000305}; Single-pass type I
membrane protein {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: GPC3_HUMAN FUNCTION: Cell surface proteoglycan that bears heparan sulfate.
Inhibits the dipeptidyl peptidase activity of DPP4. May be
involved in the suppression/modulation of growth in the
predominantly mesodermal tissues and organs. May play a role in
the modulation of IGF2 interactions with its receptor and thereby
modulate its function. May regulate growth and tumor
predisposition. {ECO:0000269|PubMed:17549790}.
SUBCELLULAR LOCATION: Cell membrane {ECO:0000250}; Lipid-anchor,
GPI-anchor {ECO:0000250}; Extracellular side {ECO:0000250}.
SUBCELLULAR LOCATION: Secreted glypican-3: Secreted, extracellular
space {ECO:0000250}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: FKB14_HUMAN FUNCTION: PPIases accelerate the folding of proteins during
protein synthesis.
SUBCELLULAR LOCATION: Endoplasmic reticulum lumen
{ECO:0000255|PROSITE-ProRule:PRU10138,
ECO:0000269|PubMed:22265013}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TXD15_HUMAN SUBCELLULAR LOCATION: Membrane {ECO:0000305}; Single-pass type I
membrane protein {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: DSC3_HUMAN FUNCTION: Component of intercellular desmosome junctions. Involved
in the interaction of plaque proteins and intermediate filaments
mediating cell-cell adhesion. May contribute to epidermal cell
positioning (stratification) by mediating differential
adhesiveness between cells that express different isoforms.
SUBCELLULAR LOCATION: Cell membrane; Single-pass type I membrane
protein. Cell junction, desmosome.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: LMA2L_HUMAN FUNCTION: May be involved in the regulation of export from the
endoplasmic reticulum of a subset of glycoproteins. May function
as a regulator of ERGIC-53. {ECO:0000269|PubMed:12878160}.
SUBCELLULAR LOCATION: Endoplasmic reticulum membrane; Single-pass
type I membrane protein. Golgi apparatus membrane; Single-pass
type I membrane protein. Note=Predominantly found in the
endoplasmic reticulum. Partly found in the Golgi.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: 1B07_HUMAN FUNCTION: Involved in the presentation of foreign antigens to the
immune system.
SUBCELLULAR LOCATION: Membrane; Single-pass type I membrane
protein.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: NIPA1_HUMAN FUNCTION: Acts as a Mg(2+) transporter. Can also transport other
divalent cations such as Fe(2+), Sr(2+), Ba(2+), Mn(2+) and Co(2+)
but to a much less extent than Mg(2+) (By similarity).
{ECO:0000250}.
SUBCELLULAR LOCATION: Cell membrane {ECO:0000305}; Multi-pass
membrane protein {ECO:0000305}. Early endosome {ECO:0000250}.
Note=Recruited to the cell membrane in response to low
extracellular magnesium. {ECO:0000250}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CNTN1_HUMAN FUNCTION: Contactins mediate cell surface interactions during
nervous system development. Involved in the formation of paranodal
axo-glial junctions in myelinated peripheral nerves and in the
signaling between axons and myelinating glial cells via its
association with CNTNAP1. Participates in oligodendrocytes
generation by acting as a ligand of NOTCH1. Its association with
NOTCH1 promotes NOTCH1 activation through the released notch
intracellular domain (NICD) and subsequent translocation to the
nucleus. Interaction with TNR induces a repulsion of neurons and
an inhibition of neurite outgrowth (By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Isoform 1: Cell membrane; Lipid-anchor, GPI-
anchor; Extracellular side.
SUBCELLULAR LOCATION: Isoform 2: Cell membrane; Lipid-anchor, GPI-
anchor; Extracellular side.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: PCDH7_HUMAN SUBCELLULAR LOCATION: Cell membrane; Single-pass type I membrane
protein.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: PCYOX_HUMAN FUNCTION: Involved in the degradation of prenylated proteins.
Cleaves the thioether bond of prenyl-L-cysteines, such as
farnesylcysteine and geranylgeranylcysteine.
SUBCELLULAR LOCATION: Lysosome.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: APOE_HUMAN FUNCTION: Mediates the binding, internalization, and catabolism of
lipoprotein particles. It can serve as a ligand for the LDL (apo
B/E) receptor and for the specific apo-E receptor (chylomicron
remnant) of hepatic tissues. {ECO:0000303|PubMed:3283935}.
SUBCELLULAR LOCATION: Secreted {ECO:0000303|PubMed:3283935}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: AMRP_HUMAN FUNCTION: Molecular chaperone for LDL receptor-related proteins
that may regulate their ligand binding activity along the
secretory pathway. {ECO:0000269|PubMed:7774585}.
SUBCELLULAR LOCATION: Rough endoplasmic reticulum lumen
{ECO:0000269|PubMed:7774585}. Endoplasmic reticulum-Golgi
intermediate compartment lumen {ECO:0000269|PubMed:7774585}. Golgi
apparatus, cis-Golgi network {ECO:0000269|PubMed:7774585}. Golgi
apparatus lumen {ECO:0000269|PubMed:7774585}. Endosome lumen
{ECO:0000269|PubMed:7774585}. Cell surface
{ECO:0000269|PubMed:11384978}. Note=May be associated with
receptors at the cell surface. {ECO:0000269|PubMed:11384978}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: PCDGK_HUMAN FUNCTION: Potential calcium-dependent cell-adhesion protein. May
be involved in the establishment and maintenance of specific
neuronal connections in the brain.
SUBCELLULAR LOCATION: Cell membrane {ECO:0000250}; Single-pass
type I membrane protein {ECO:0000250}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: LMAN2_HUMAN FUNCTION: Plays a role as an intracellular lectin in the early
secretory pathway. Interacts with N-acetyl-D-galactosamine and
high-mannose type glycans and may also bind to O-linked glycans.
Involved in the transport and sorting of glycoproteins carrying
high mannose-type glycans (By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Endoplasmic reticulum-Golgi intermediate
compartment membrane {ECO:0000269|PubMed:10444376}; Single-pass
type I membrane protein {ECO:0000269|PubMed:10444376}. Golgi
apparatus membrane {ECO:0000269|PubMed:10444376}; Single-pass
membrane protein {ECO:0000269|PubMed:10444376}. Endoplasmic
reticulum membrane {ECO:0000269|PubMed:10444376}; Single-pass type
I membrane protein {ECO:0000269|PubMed:10444376}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: KDEL2_HUMAN SUBCELLULAR LOCATION: Endoplasmic reticulum lumen
{ECO:0000255|PROSITE-ProRule:PRU10138}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: GPC6_HUMAN FUNCTION: Cell surface proteoglycan that bears heparan sulfate.
Putative cell surface coreceptor for growth factors, extracellular
matrix proteins, proteases and anti-proteases (By similarity).
Enhances migration and invasion of cancer cells through WNT5A
signaling. {ECO:0000250, ECO:0000269|PubMed:21871017}.
SUBCELLULAR LOCATION: Cell membrane {ECO:0000250}; Lipid-anchor,
GPI-anchor {ECO:0000250}; Extracellular side {ECO:0000250}.
SUBCELLULAR LOCATION: Secreted glypican-6: Secreted, extracellular
space {ECO:0000250}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TECT3_HUMAN FUNCTION: Part of the tectonic-like complex which is required for
tissue-specific ciliogenesis and may regulate ciliary membrane
composition (By similarity). May be involved in apoptosis
regulation. Necessary for signal transduction through the sonic
hedgehog (Shh) signaling pathway. {ECO:0000250,
ECO:0000269|PubMed:17464193, ECO:0000269|PubMed:22883145}.
SUBCELLULAR LOCATION: Membrane {ECO:0000305}; Single-pass type I
membrane protein {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TMX3_HUMAN FUNCTION: Probable disulfide isomerase, which participates in the
folding of proteins containing disulfide bonds. May act as a
dithiol oxidase. {ECO:0000269|PubMed:15623505}.
SUBCELLULAR LOCATION: Endoplasmic reticulum membrane
{ECO:0000269|PubMed:15623505}; Single-pass membrane protein
{ECO:0000269|PubMed:15623505}.
CLEAVAGE EVENTS OBSERVED:Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' RPN1_HUMAN SSEAPPLINEDVK 25 PGSA | SSEA RPN1_HUMAN SSEAPPLINEDVKR 25 PGSA | SSEA RPN1_HUMAN SASSEAPPLINEDVKR 23 PAPG | SASS 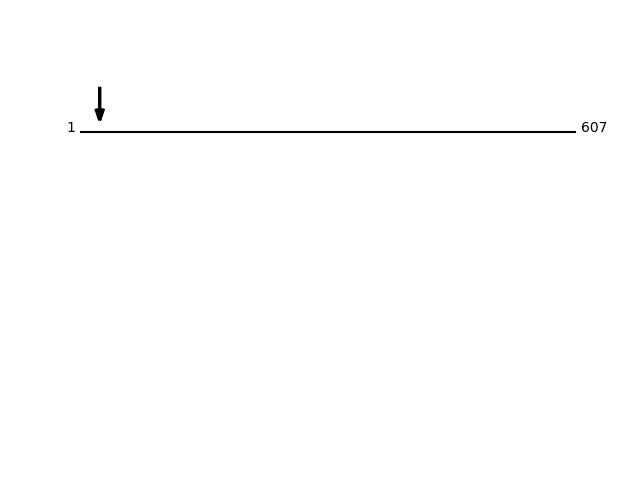
P19022 - Cadherin-2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CADH2_HUMAN TFYGEVPENR 386 FTAM | TFYG CADH2_HUMAN TVAAENQVPLAK 461 MFVL | TVAA CADH2_HUMAN SLKPTLTEESVK 122 AVKL | SLKP CADH2_HUMAN TFTAQDPDR 518 TMLT | TFTA CADH2_HUMAN FYGEVPENR 387 TAMT | FYGE CADH2_HUMAN TAQDPDRYMQQNIR 520 LTTF | TAQD 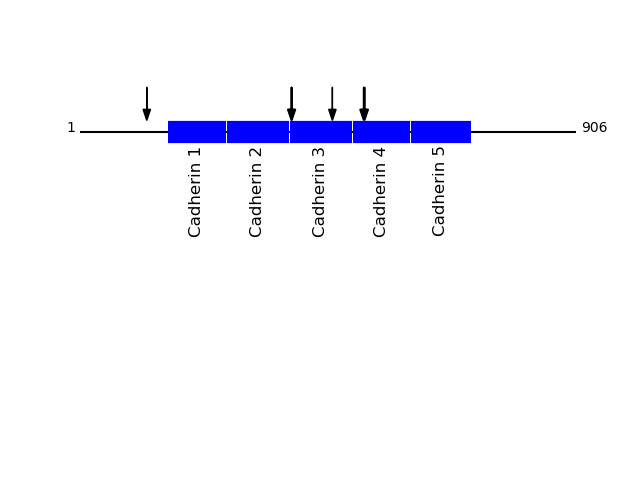
Q8NBS9 - Thioredoxin domain-containing protein 5Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TXND5_HUMAN AAADGPPAADGEDGQDPHSK 44 EAAA | AAAD TXND5_HUMAN AADGPPAADGEDGQDPHSK 45 AAAA | AADG 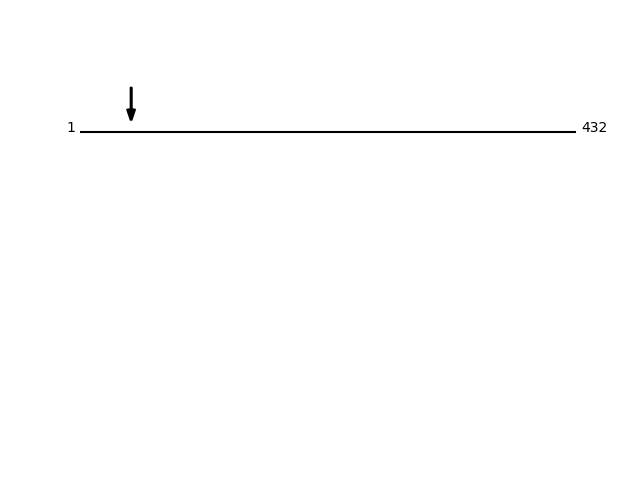
Q99805 - Transmembrane 9 superfamily member 2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TM9S2_HUMAN SVSFEEDDKIR 272 AYTY | SVSF 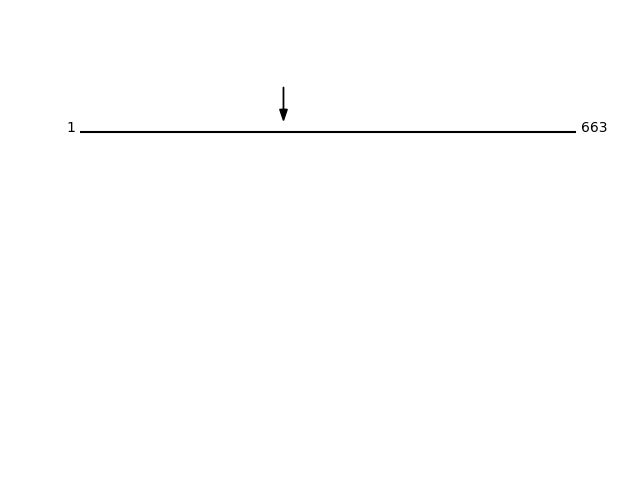
Q8N129 - Protein canopy homolog 4Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CNPY4_HUMAN GMLKEEDDDTER 22 EAWA | GMLK CNPY4_HUMAN GMLKEEDDDTERLPSK 22 EAWA | GMLK 
Q9ULF5 - Zinc transporter ZIP10Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' S39AA_HUMAN HEEHDHGPEALHR 26 CNHC | HEEH S39AA_HUMAN GMTELEPSK 42 RQHR | GMTE S39AA_HUMAN GMTELEPSK 42 RQHR | GMTE 
Q9H1E5 - Thioredoxin-related transmembrane protein 4Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TMX4_HUMAN TAGPEEAALPPEQSR 24 AVAA | TAGP 
Q92504 - Zinc transporter SLC39A7Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' S39A7_HUMAN GYSHESLYHR 95 GHSH | GYSH S39A7_HUMAN SHESLYHR 97 SHGY | SHES 
P14314 - Glucosidase 2 subunit betaProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' GLU2B_HUMAN AQQEQELAADAFK 207 AAAK | AQQE GLU2B_HUMAN SLEDQVEMLR 168 AGKK | SLED 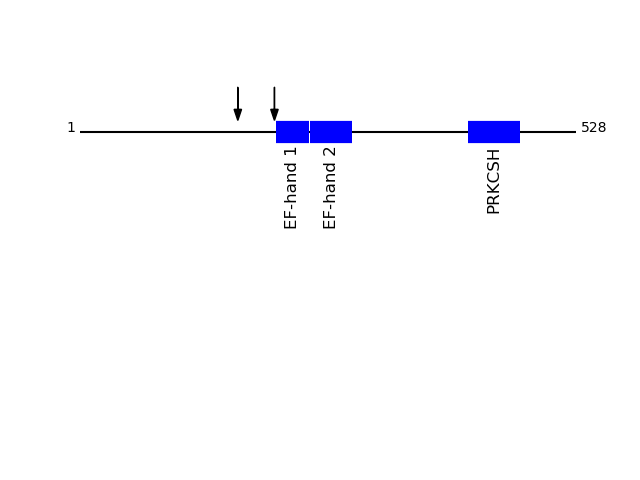
Q92643 - GPI-anchor transamidaseProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' GPI8_HUMAN SHIEDQAEQFFR 28 SVAA | SHIE 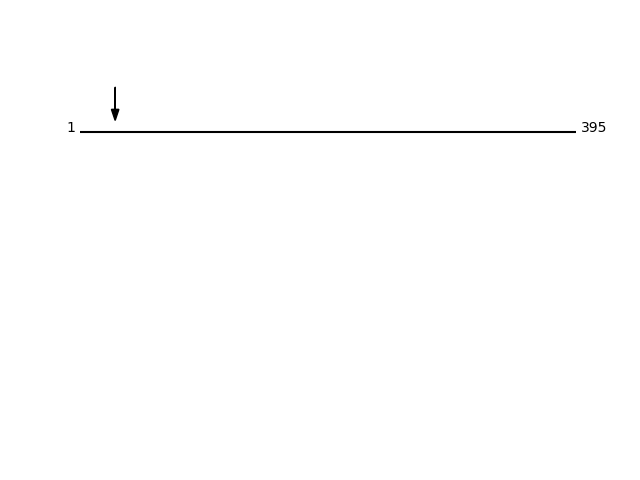
O94898 - Leucine-rich repeats and immunoglobulin-like domains protein 2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' LRIG2_HUMAN GLCPAPCSCR 41 AAGA | GLCP 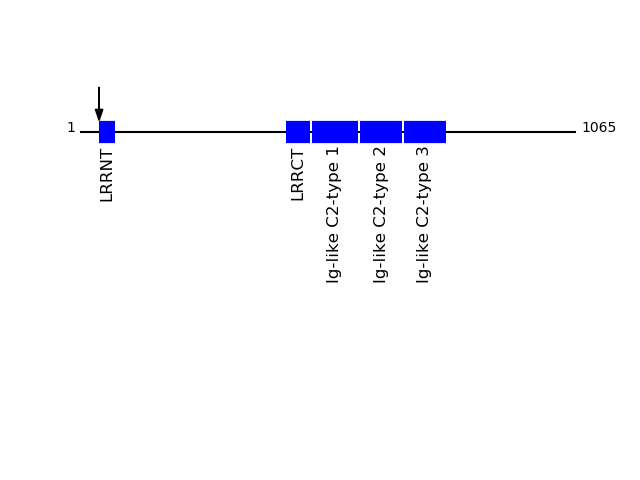
O14763 - Tumor necrosis factor receptor superfamily member 10BProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TR10B_HUMAN ALITQQDLAPQQR 54 SAES | ALIT 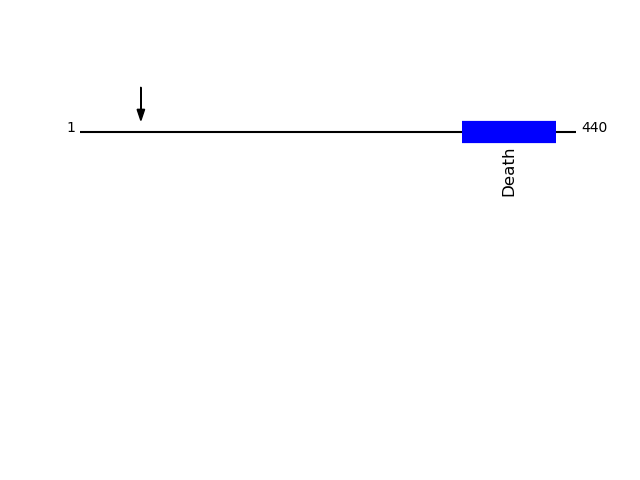
P62341 - Selenoprotein T {ECO:0000303|PubMed:27645994}Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' SELT_HUMAN SANLGGVPSK 20 RSEA | SANL 
Q04721 - Neurogenic locus notch homolog protein 2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' NOTC2_HUMAN SLPGEQEQEVAGSK 1609 MTRR | SLPG 
Q9Y624 - Junctional adhesion molecule AProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' JAM1_HUMAN SVTVHSSEPEVR 28 LALG | SVTV 
Q9NPA0 - ER membrane protein complex subunit 7Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' EMC7_HUMAN SEVPGAAAEGSGGSGVGIGDR 24 DVQS | SEVP 
Q5JRA6 - Transport and Golgi organization protein 1 homolog {ECO:0000305}Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TGO1_HUMAN GLAGEPEGELSKEDHENTEK 866 LKTS | GLAG 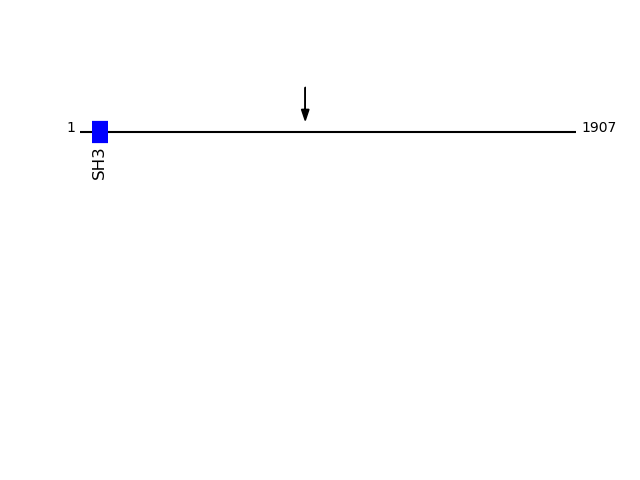
P18827 - Syndecan-1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' SDC1_HUMAN STSTLPAGEGPK 89 ATAA | STST 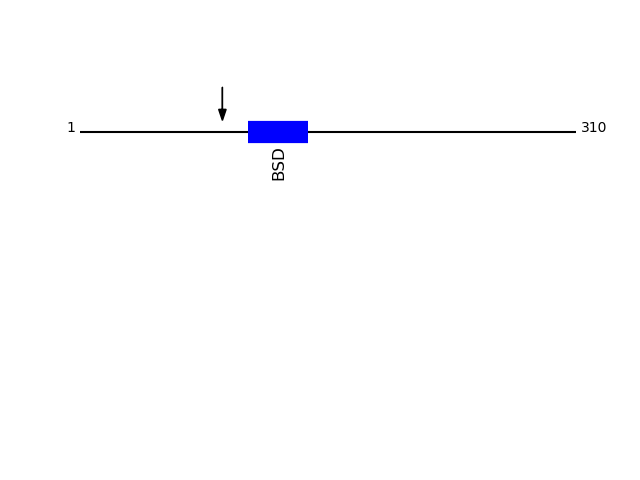
Q13586 - Stromal interaction molecule 1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' STIM1_HUMAN SSGANSEESTAAEFCR 35 ATGT | SSGA 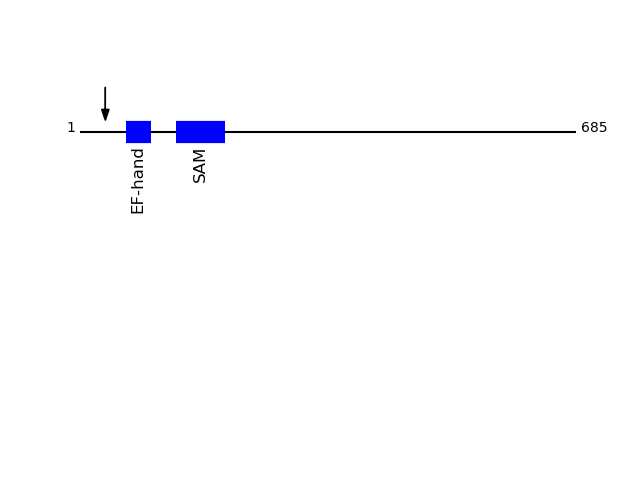
Q16625 - OccludinProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' OCLN_HUMAN GYGYGYGGYTDPR 123 GYGY | GYGY 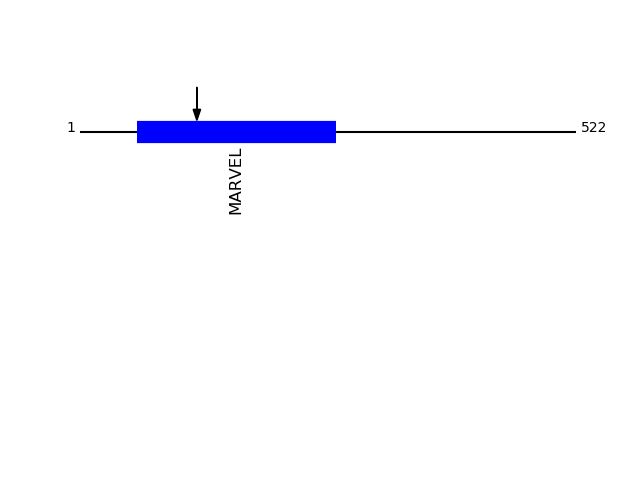
P14209 - CD99 antigenProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CD99_HUMAN SFSDADLADGVSGGEGK 89 SSSG | SFSD 
P51654 - Glypican-3Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' GPC3_HUMAN HVEHEETLSSR 377 LKVA | HVEH 
Q9NWM8 - Peptidyl-prolyl cis-trans isomerase FKBP14Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' FKB14_HUMAN ALIPEPEVK 20 SLIG | ALIP 
Q96J42 - Thioredoxin domain-containing protein 15Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TXD15_HUMAN SVIPGEAEDK 95 MVML | SVIP 
Q14574 - Desmocollin-3Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' DSC3_HUMAN TADGYSADLPLPLPIR 217 AYAS | TADG 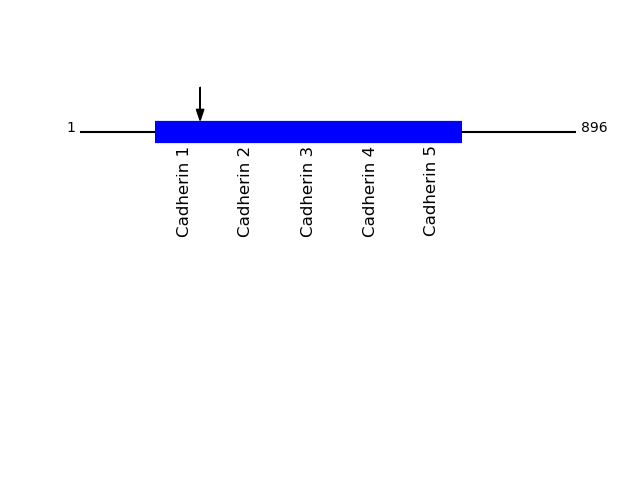
Q9H0V9 - VIP36-like proteinProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' LMA2L_HUMAN GQTFEYLK 45 QVGA | GQTF LMA2L_HUMAN GQTFEYLKR 45 QVGA | GQTF 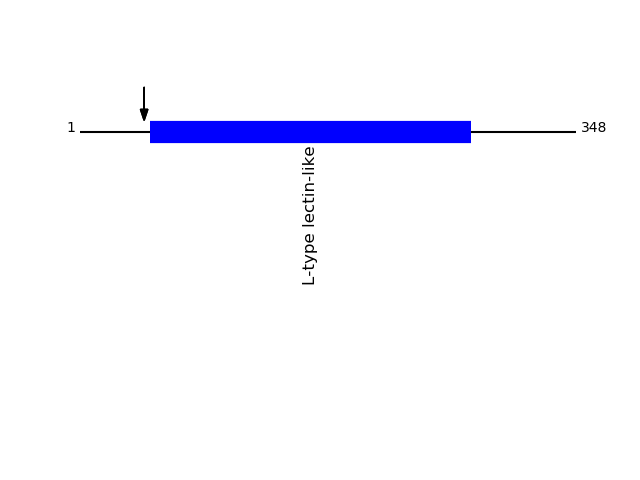
P01889 - HLA class I histocompatibility antigen, B-7 alpha chainProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' 1B07_HUMAN GYVDDTQFVR 50 FISV | GYVD 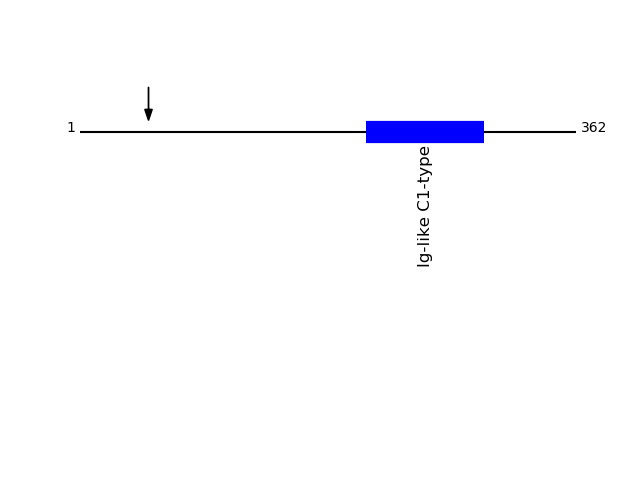
Q7RTP0 - Magnesium transporter NIPA1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' NIPA1_HUMAN AAAAAAAAAGEGAR 8 AAAA | AAAA 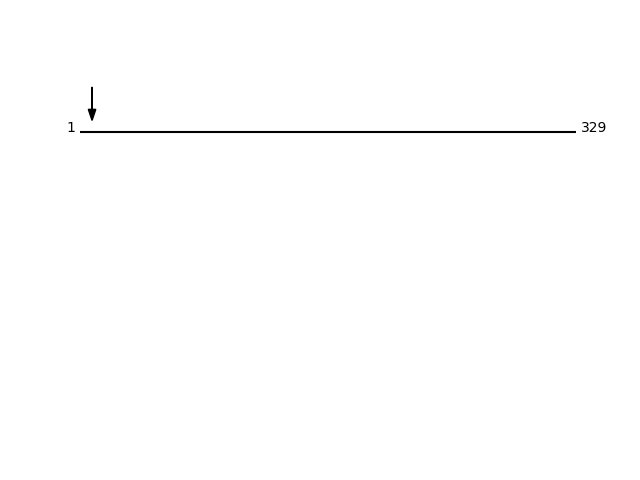
Q12860 - Contactin-1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CNTN1_HUMAN SAQDAPSEAPTEVGVK 803 AVIN | SAQD 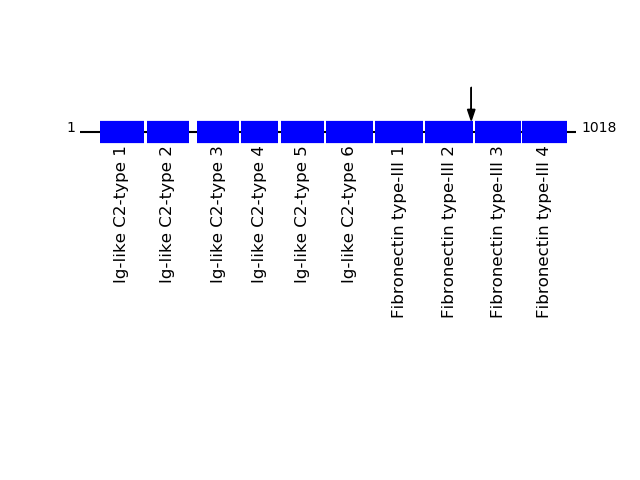
O60245 - Protocadherin-7Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' PCDH7_HUMAN HIPLTQDIAGDPSYEISK 858 ARSL | HIPL PCDH7_HUMAN AVDGGDPPR 714 FRVK | AVDG 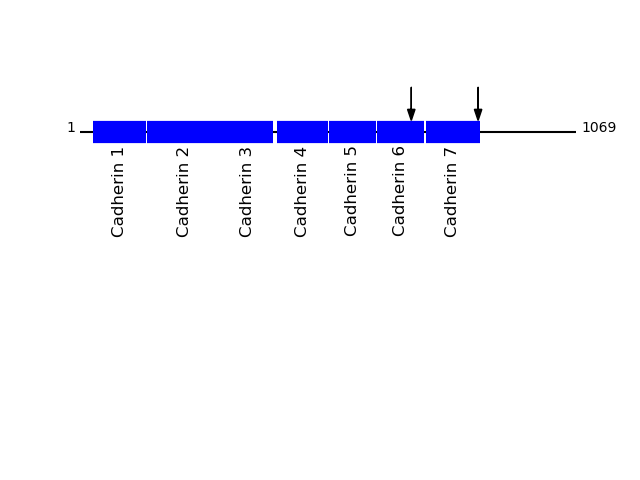
Q9UHG3 - Prenylcysteine oxidase 1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' PCYOX_HUMAN GCPEGAELR 23 LCSC | GCPE 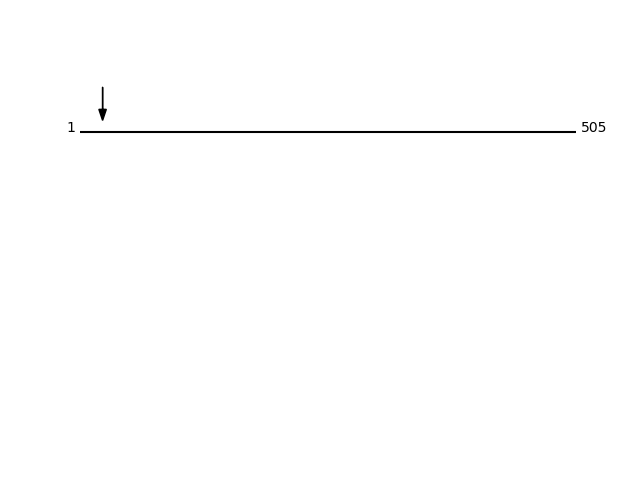
P02649 - Apolipoprotein EProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' APOE_HUMAN TVGSLAGQPLQER 212 VRAA | TVGS 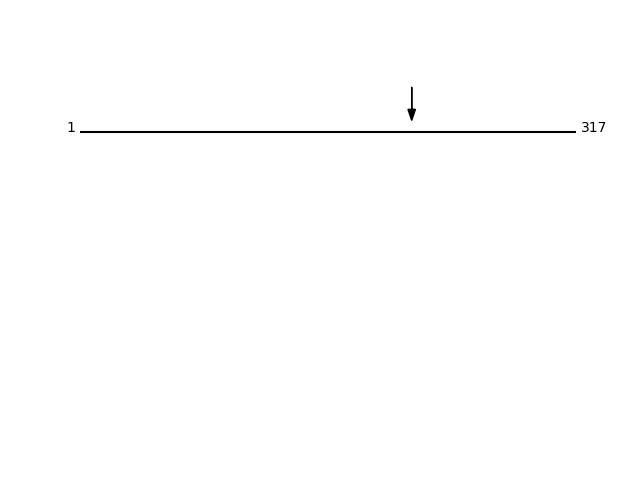
P30533 - Alpha-2-macroglobulin receptor-associated proteinProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' AMRP_HUMAN SHQGYSTEAEFEEPR 242 LRRV | SHQG 
Q9UN70 - Protocadherin gamma-C3Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' PCDGK_HUMAN STVIHYEIPEER 30 LNKA | STVI 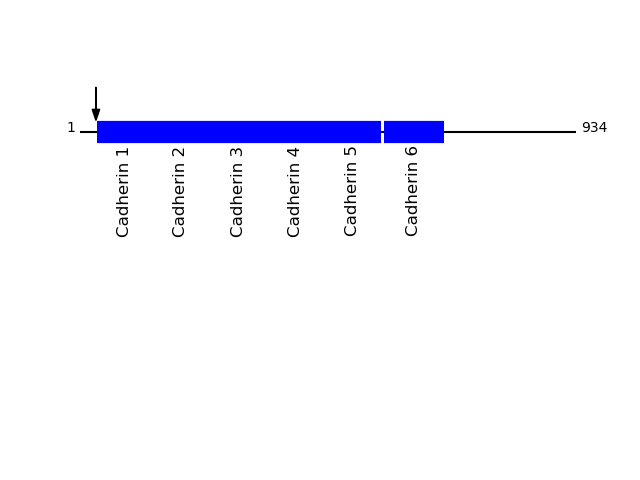
Q12907 - Vesicular integral-membrane protein VIP36Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' LMAN2_HUMAN DITDGNSEHLK 45 SVTA | DITD 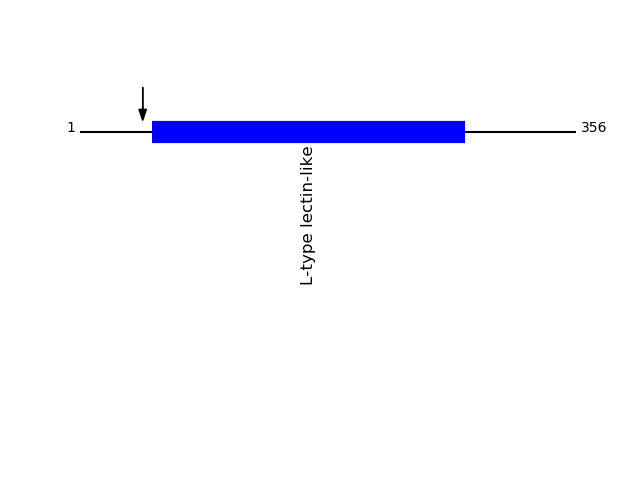
Q7Z4H8 - KDEL motif-containing protein 2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' KDEL2_HUMAN GAPEVLVSAPR 21 LVAA | GAPE 
Q9Y625 - Glypican-6Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' GPC6_HUMAN SAPENFNTR 355 RSAR | SAPE 
Q6NUS6 - Tectonic-3Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TECT3_HUMAN STAASLTSPR 370 AFQQ | STAA 
Q96JJ7 - Protein disulfide-isomerase TMX3