Listed below are N-terminal peptides that were captured and enriched from the surface of a HEK293T cell line expressing subtiligase-Y217K-TM.
GRP78_HUMAN
P11021 - 78 kDa glucose-regulated protein
FUNCTION: Probably plays a role in facilitating the assembly of multimeric protein complexes inside the endoplasmic reticulum. Involved in the correct folding of proteins and degradation of misfolded proteins via its interaction with DNAJC10, probably to facilitate the release of DNAJC10 from its substrate. {ECO:0000269|PubMed:2294010, ECO:0000269|PubMed:23769672, ECO:0000269|PubMed:23990668}.
SUBCELLULAR LOCATION: Endoplasmic reticulum lumen {ECO:0000269|PubMed:21080038, ECO:0000269|PubMed:21289099, ECO:0000269|PubMed:23990668}. Melanosome. Cytoplasm {ECO:0000250}. Note=Identified by mass spectrometry in melanosome fractions from stage I to stage IV.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: RPN1_HUMAN FUNCTION: Essential subunit of the N-oligosaccharyl transferase
(OST) complex which catalyzes the transfer of a high mannose
oligosaccharide from a lipid-linked oligosaccharide donor to an
asparagine residue within an Asn-X-Ser/Thr consensus motif in
nascent polypeptide chains. {ECO:0000250|UniProtKB:E2RQ08,
ECO:0000250|UniProtKB:Q9GMB0, ECO:0000305}.
SUBCELLULAR LOCATION: Endoplasmic reticulum
{ECO:0000250|UniProtKB:E2RQ08, ECO:0000250|UniProtKB:Q9GMB0}.
Endoplasmic reticulum membrane; Single-pass type I membrane
protein {ECO:0000305}. Melanosome. Note=Identified by mass
spectrometry in melanosome fractions from stage I to stage IV.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CADH2_HUMAN FUNCTION: Cadherins are calcium-dependent cell adhesion proteins.
They preferentially interact with themselves in a homophilic
manner in connecting cells; cadherins may thus contribute to the
sorting of heterogeneous cell types. Acts as a regulator of neural
stem cells quiescence by mediating anchorage of neural stem cells
to ependymocytes in the adult subependymal zone: upon cleavage by
MMP24, CDH2-mediated anchorage is affected, leading to modulate
neural stem cell quiescence. CDH2 may be involved in neuronal
recognition mechanism. In hippocampal neurons, may regulate
dendritic spine density (By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Cell membrane
{ECO:0000250|UniProtKB:P15116}; Single-pass type I membrane
protein {ECO:0000255}. Cell membrane, sarcolemma
{ECO:0000250|UniProtKB:P15116}. Cell junction
{ECO:0000269|PubMed:28169360}. Cell surface
{ECO:0000250|UniProtKB:P15116}. Note=Colocalizes with TMEM65 at
the intercalated disk in cardiomyocytes. Colocalizes with OBSCN at
the intercalated disk and at sarcolemma in cardiomyocytes.
{ECO:0000250|UniProtKB:P15116}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CALU_HUMAN FUNCTION: Involved in regulation of vitamin K-dependent
carboxylation of multiple N-terminal glutamate residues. Seems to
inhibit gamma-carboxylase GGCX. Binds 7 calcium ions with a low
affinity (By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Endoplasmic reticulum membrane
{ECO:0000269|PubMed:10222138}. Golgi apparatus
{ECO:0000269|PubMed:10222138}. Secreted {ECO:0000305}. Melanosome
{ECO:0000269|PubMed:12643545}. Sarcoplasmic reticulum lumen
{ECO:0000305}. Note=Identified by mass spectrometry in melanosome
fractions from stage I to stage IV. {ECO:0000269|PubMed:12643545}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TGO1_HUMAN FUNCTION: Plays a role in the transport of cargos that are too
large to fit into COPII-coated vesicles and require specific
mechanisms to be incorporated into membrane-bound carriers and
exported from the endoplasmic reticulum. This protein is required
for collagen VII (COL7A1) secretion by loading COL7A1 into
transport carriers. It may participate in cargo loading of COL7A1
at endoplasmic reticulum exit sites by binding to COPII coat
subunits Sec23/24 and guiding SH3-bound COL7A1 into a growing
carrier. Does not play a role in global protein secretion and is
apparently specific to COL7A1 cargo loading. However, it may
participate in secretion of other proteins in cells that do not
secrete COL7A1. It is also specifically required for the secretion
of lipoproteins by participating in their export from the
endoplasmic reticulum (PubMed:27138255).
{ECO:0000269|PubMed:19269366, ECO:0000269|PubMed:27138255}.
SUBCELLULAR LOCATION: Endoplasmic reticulum membrane
{ECO:0000269|PubMed:19269366, ECO:0000269|PubMed:21525241};
Single-pass membrane protein {ECO:0000269|PubMed:19269366}.
Note=Localizes at endoplasmic reticulum exit sites. After loading
of COL7A1 into transport carriers, it is not incorporated into
COPII carriers and remains in the endoplasmic reticulum membrane.
{ECO:0000269|PubMed:19269366, ECO:0000269|PubMed:21525241}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: LMAN2_HUMAN FUNCTION: Plays a role as an intracellular lectin in the early
secretory pathway. Interacts with N-acetyl-D-galactosamine and
high-mannose type glycans and may also bind to O-linked glycans.
Involved in the transport and sorting of glycoproteins carrying
high mannose-type glycans (By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Endoplasmic reticulum-Golgi intermediate
compartment membrane {ECO:0000269|PubMed:10444376}; Single-pass
type I membrane protein {ECO:0000269|PubMed:10444376}. Golgi
apparatus membrane {ECO:0000269|PubMed:10444376}; Single-pass
membrane protein {ECO:0000269|PubMed:10444376}. Endoplasmic
reticulum membrane {ECO:0000269|PubMed:10444376}; Single-pass type
I membrane protein {ECO:0000269|PubMed:10444376}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TXND5_HUMAN FUNCTION: Possesses thioredoxin activity. Has been shown to reduce
insulin disulfide bonds. Also complements protein disulfide-
isomerase deficiency in yeast (By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Endoplasmic reticulum lumen
{ECO:0000255|PROSITE-ProRule:PRU10138}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TM9S2_HUMAN FUNCTION: In the intracellular compartments, may function as a
channel or small molecule transporter.
SUBCELLULAR LOCATION: Endosome membrane {ECO:0000305}; Multi-pass
membrane protein {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: PDIA4_HUMAN SUBCELLULAR LOCATION: Endoplasmic reticulum lumen
{ECO:0000269|PubMed:12643545}. Melanosome
{ECO:0000269|PubMed:17081065}. Note=Identified by mass
spectrometry in melanosome fractions from stage I to stage IV
(PubMed:17081065). {ECO:0000269|PubMed:17081065}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: 1B07_HUMAN FUNCTION: Involved in the presentation of foreign antigens to the
immune system.
SUBCELLULAR LOCATION: Membrane; Single-pass type I membrane
protein.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: DSG2_HUMAN FUNCTION: Component of intercellular desmosome junctions. Involved
in the interaction of plaque proteins and intermediate filaments
mediating cell-cell adhesion.
SUBCELLULAR LOCATION: Cell membrane; Single-pass type I membrane
protein. Cell junction, desmosome.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: S39AA_HUMAN FUNCTION: May act as a zinc-influx transporter.
{ECO:0000269|PubMed:17359283}.
SUBCELLULAR LOCATION: Membrane {ECO:0000305}; Multi-pass membrane
protein {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: GLU2B_HUMAN FUNCTION: Regulatory subunit of glucosidase II.
{ECO:0000269|PubMed:10929008}.
SUBCELLULAR LOCATION: Endoplasmic reticulum {ECO:0000255|PROSITE-
ProRule:PRU10138}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: NUCB2_HUMAN FUNCTION: Calcium-binding protein. May have a role in calcium
homeostasis.
FUNCTION: Nesfatin-1: Anorexigenic peptide, seems to play an
important role in hypothalamic pathways regulating food intake and
energy homeostasis, acting in a leptin-independent manner. May
also exert hypertensive roles and modulate blood pressure through
directly acting on peripheral arterial resistance (By similarity).
{ECO:0000250}.
SUBCELLULAR LOCATION: Golgi apparatus
{ECO:0000269|PubMed:11749975}. Membrane
{ECO:0000269|PubMed:11749975}; Peripheral membrane protein
{ECO:0000269|PubMed:11749975}. Cytoplasm
{ECO:0000269|PubMed:11749975}. Secreted
{ECO:0000269|PubMed:11749975}. Endoplasmic reticulum
{ECO:0000250}. Nucleus envelope {ECO:0000250}. Note=Golgi
retention is mediated by its N-terminal region.
SUBCELLULAR LOCATION: Nesfatin-1: Secreted.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: PDIA1_HUMAN FUNCTION: This multifunctional protein catalyzes the formation,
breakage and rearrangement of disulfide bonds. At the cell
surface, seems to act as a reductase that cleaves disulfide bonds
of proteins attached to the cell. May therefore cause structural
modifications of exofacial proteins. Inside the cell, seems to
form/rearrange disulfide bonds of nascent proteins. At high
concentrations, functions as a chaperone that inhibits aggregation
of misfolded proteins. At low concentrations, facilitates
aggregation (anti-chaperone activity). May be involved with other
chaperones in the structural modification of the TG precursor in
hormone biogenesis. Also acts a structural subunit of various
enzymes such as prolyl 4-hydroxylase and microsomal
triacylglycerol transfer protein MTTP. Receptor for LGALS9; the
interaction retains P4HB at the cell surface of Th2 T helper
cells, increasing disulfide reductase activity at the plasma
membrane, altering the plasma membrane redox state and enhancing
cell migration (PubMed:21670307). {ECO:0000269|PubMed:10636893,
ECO:0000269|PubMed:12485997, ECO:0000269|PubMed:21670307}.
SUBCELLULAR LOCATION: Endoplasmic reticulum
{ECO:0000269|PubMed:23475612}. Endoplasmic reticulum lumen
{ECO:0000269|PubMed:10636893, ECO:0000269|PubMed:23475612}.
Melanosome {ECO:0000269|PubMed:12643545,
ECO:0000269|PubMed:17081065}. Cell membrane
{ECO:0000269|PubMed:21670307}; Peripheral membrane protein
{ECO:0000305}. Note=Highly abundant. In some cell types, seems to
be also secreted or associated with the plasma membrane, where it
undergoes constant shedding and replacement from intracellular
sources (Probable). Localizes near CD4-enriched regions on
lymphoid cell surfaces (PubMed:11181151). Identified by mass
spectrometry in melanosome fractions from stage I to stage IV
(PubMed:10636893). Colocalizes with MTTP in the endoplasmic
reticulum (PubMed:23475612). {ECO:0000269|PubMed:10636893,
ECO:0000269|PubMed:11181151, ECO:0000269|PubMed:23475612,
ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: RCN1_HUMAN FUNCTION: May regulate calcium-dependent activities in the
endoplasmic reticulum lumen or post-ER compartment.
SUBCELLULAR LOCATION: Endoplasmic reticulum lumen.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: RCN2_HUMAN FUNCTION: Not known. Binds calcium.
SUBCELLULAR LOCATION: Endoplasmic reticulum lumen.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: JAM1_HUMAN FUNCTION: Seems to play a role in epithelial tight junction
formation. Appears early in primordial forms of cell junctions and
recruits PARD3 (PubMed:11489913). The association of the PARD6-
PARD3 complex may prevent the interaction of PARD3 with JAM1,
thereby preventing tight junction assembly (By similarity). Plays
a role in regulating monocyte transmigration involved in integrity
of epithelial barrier (By similarity). Ligand for integrin alpha-
L/beta-2 involved in memory T-cell and neutrophil transmigration
(PubMed:11812992). Involved in platelet activation
(PubMed:10753840). {ECO:0000250|UniProtKB:O88792,
ECO:0000269|PubMed:10753840, ECO:0000269|PubMed:11489913,
ECO:0000269|PubMed:11812992}.
FUNCTION: (Microbial infection) Acts as a receptor for Mammalian
reovirus sigma-1. {ECO:0000269|PubMed:11239401}.
FUNCTION: (Microbial infection) Acts as a receptor for Human
Rotavirus strain Wa. {ECO:0000269|PubMed:25481868}.
SUBCELLULAR LOCATION: Cell junction, tight junction
{ECO:0000269|PubMed:11171323}. Cell membrane
{ECO:0000269|PubMed:11171323}; Single-pass type I membrane protein
{ECO:0000269|PubMed:11171323}. Note=Localized at tight junctions
of both epithelial and endothelial cells.
{ECO:0000269|PubMed:11171323}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TMX4_HUMAN SUBCELLULAR LOCATION: Membrane {ECO:0000305}; Single-pass type I
membrane protein {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: NOTC2_HUMAN FUNCTION: Functions as a receptor for membrane-bound ligands
Jagged1, Jagged2 and Delta1 to regulate cell-fate determination.
Upon ligand activation through the released notch intracellular
domain (NICD) it forms a transcriptional activator complex with
RBPJ/RBPSUH and activates genes of the enhancer of split locus.
Affects the implementation of differentiation, proliferation and
apoptotic programs (By similarity). Involved in bone remodeling
and homeostasis. In collaboration with RELA/p65 enhances NFATc1
promoter activity and positively regulates RANKL-induced
osteoclast differentiation. Positively regulates self-renewal of
liver cancer cells (PubMed:25985737).
{ECO:0000250|UniProtKB:O35516, ECO:0000269|PubMed:21378985,
ECO:0000269|PubMed:21378989, ECO:0000269|PubMed:25985737}.
SUBCELLULAR LOCATION: Notch 2 extracellular truncation: Cell
membrane {ECO:0000269|PubMed:9244302}; Single-pass type I membrane
protein {ECO:0000269|PubMed:9244302}.
SUBCELLULAR LOCATION: Notch 2 intracellular domain: Nucleus
{ECO:0000269|PubMed:25985737}. Cytoplasm
{ECO:0000269|PubMed:25985737}. Note=Following proteolytical
processing NICD is translocated to the nucleus. Retained at the
cytoplasm by C8orf4 (PubMed:25985737).
{ECO:0000269|PubMed:25985737}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: SUN1_HUMAN FUNCTION: As a component of the LINC (LInker of Nucleoskeleton and
Cytoskeleton) complex involved in the connection between the
nuclear lamina and the cytoskeleton. The nucleocytoplasmic
interactions established by the LINC complex play an important
role in the transmission of mechanical forces across the nuclear
envelope and in nuclear movement and positioning. Required for
interkinetic nuclear migration (INM) and essential for
nucleokinesis and centrosome-nucleus coupling during radial
neuronal migration in the cerebral cortex and during glial
migration. Involved in telomere attachment to nuclear envelope in
the prophase of meiosis implicating a SUN1/2:KASH5 LINC complex in
which SUN1 and SUN2 seem to act at least partial redundantly.
Required for gametogenesis and involved in selective gene
expression of coding and non-coding RNAs needed for gametogenesis.
Helps to define the distribution of nuclear pore complexes (NPCs).
Required for efficient localization of SYNE4 in the nuclear
envelope. May be involved in nuclear remodeling during sperm head
formation in spermatogenenis. {ECO:0000250|UniProtKB:Q9D666,
ECO:0000269|PubMed:18039933, ECO:0000269|PubMed:18396275}.
SUBCELLULAR LOCATION: Nucleus inner membrane
{ECO:0000269|PubMed:12958361, ECO:0000269|PubMed:16445915,
ECO:0000269|PubMed:17132086, ECO:0000269|PubMed:18845190,
ECO:0000269|PubMed:19933576}; Single-pass type II membrane protein
{ECO:0000269|PubMed:12958361, ECO:0000269|PubMed:16445915,
ECO:0000269|PubMed:17132086, ECO:0000269|PubMed:18845190,
ECO:0000269|PubMed:19933576}. Note=At oocyte MI stage localized
around the spindle, at MII stage localized to the spindle poles.
{ECO:0000250|UniProtKB:Q9D666}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: S39A7_HUMAN FUNCTION: Zinc transporter, that transports Zn(2+) from the
endoplasmic reticulum/Golgi apparatus to the cytosol. Transport is
stimulated by growth factors, such as EGF, and Ca(2+), as well as
by exogenous Zn(2+). {ECO:0000269|PubMed:14525538,
ECO:0000269|PubMed:15705588, ECO:0000269|PubMed:22317921}.
SUBCELLULAR LOCATION: Endoplasmic reticulum membrane; Multi-pass
membrane protein. Golgi apparatus, cis-Golgi network membrane.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: MESD_HUMAN FUNCTION: Chaperone specifically assisting the folding of beta-
propeller/EGF modules within the family of low-density lipoprotein
receptors (LDLRs). Acts as a modulator of the Wnt pathway through
chaperoning the coreceptors of the canonical Wnt pathway, LRP5 and
LRP6, to the plasma membrane. Essential for specification of
embryonic polarity and mesoderm induction.
{ECO:0000269|PubMed:15014448, ECO:0000269|PubMed:17488095}.
SUBCELLULAR LOCATION: Endoplasmic reticulum
{ECO:0000269|PubMed:15014448, ECO:0000269|PubMed:17488095}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: MCFD2_HUMAN FUNCTION: The MCFD2-LMAN1 complex forms a specific cargo receptor
for the ER-to-Golgi transport of selected proteins. Plays a role
in the secretion of coagulation factors.
{ECO:0000269|PubMed:12717434}.
SUBCELLULAR LOCATION: Endoplasmic reticulum-Golgi intermediate
compartment {ECO:0000269|PubMed:12717434}. Endoplasmic reticulum
{ECO:0000269|PubMed:12717434}. Golgi apparatus
{ECO:0000269|PubMed:12717434}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: TM2D3_HUMAN SUBCELLULAR LOCATION: Membrane {ECO:0000305}; Multi-pass membrane
protein {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CD99_HUMAN FUNCTION: Involved in T-cell adhesion processes and in spontaneous
rosette formation with erythrocytes. Plays a role in a late step
of leukocyte extravasation helping leukocytes to overcome the
endothelial basement membrane. Acts at the same site as, but
independently of, PECAM1. Involved in T-cell adhesion processes
(By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Membrane {ECO:0000305}; Single-pass type I
membrane protein {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: LMA2L_HUMAN FUNCTION: May be involved in the regulation of export from the
endoplasmic reticulum of a subset of glycoproteins. May function
as a regulator of ERGIC-53. {ECO:0000269|PubMed:12878160}.
SUBCELLULAR LOCATION: Endoplasmic reticulum membrane; Single-pass
type I membrane protein. Golgi apparatus membrane; Single-pass
type I membrane protein. Note=Predominantly found in the
endoplasmic reticulum. Partly found in the Golgi.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CNPY4_HUMAN FUNCTION: Plays a role in the regulation of the cell surface
expression of TLR4. {ECO:0000269|PubMed:16338228}.
SUBCELLULAR LOCATION: Secreted {ECO:0000305}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: ENPL_HUMAN FUNCTION: Molecular chaperone that functions in the processing and
transport of secreted proteins. When associated with CNPY3,
required for proper folding of Toll-like receptors (By
similarity). Functions in endoplasmic reticulum associated
degradation (ERAD). Has ATPase activity. {ECO:0000250,
ECO:0000269|PubMed:18264092}.
SUBCELLULAR LOCATION: Endoplasmic reticulum lumen. Melanosome.
Note=Identified by mass spectrometry in melanosome fractions from
stage I to stage IV.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CKAP4_HUMAN FUNCTION: High-affinity epithelial cell surface receptor for APF.
FUNCTION: Mediates the anchoring of the endoplasmic reticulum to
microtubules.
SUBCELLULAR LOCATION: Endoplasmic reticulum membrane; Single-pass
type II membrane protein. Cell membrane; Single-pass type II
membrane protein. Cytoplasm, cytoskeleton. Cytoplasm, perinuclear
region. Note=Translocates to the perinuclear region upon APF-
stimulation.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: PRDX4_HUMAN FUNCTION: Thiol-specific peroxidase that catalyzes the reduction
of hydrogen peroxide and organic hydroperoxides to water and
alcohols, respectively. Plays a role in cell protection against
oxidative stress by detoxifying peroxides and as sensor of
hydrogen peroxide-mediated signaling events. Regulates the
activation of NF-kappa-B in the cytosol by a modulation of I-
kappa-B-alpha phosphorylation. {ECO:0000269|PubMed:9388242}.
SUBCELLULAR LOCATION: Cytoplasm {ECO:0000269|PubMed:18052930,
ECO:0000269|PubMed:9388242}. Endoplasmic reticulum
{ECO:0000269|PubMed:18052930}. Note=Cotranslationally translocated
to and retained within the endoplasmic reticulum. A small fraction
of the protein is cytoplasmic. {ECO:0000269|PubMed:18052930}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: RECK_HUMAN FUNCTION: Negatively regulates matrix metalloproteinase-9 (MMP-9)
by suppressing MMP-9 secretion and by direct inhibition of its
enzymatic activity. RECK down-regulation by oncogenic signals may
facilitate tumor invasion and metastasis. Appears to also regulate
MMP-2 and MT1-MMP, which are involved in cancer progression.
SUBCELLULAR LOCATION: Cell membrane; Lipid-anchor, GPI-anchor.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CNTN1_HUMAN FUNCTION: Contactins mediate cell surface interactions during
nervous system development. Involved in the formation of paranodal
axo-glial junctions in myelinated peripheral nerves and in the
signaling between axons and myelinating glial cells via its
association with CNTNAP1. Participates in oligodendrocytes
generation by acting as a ligand of NOTCH1. Its association with
NOTCH1 promotes NOTCH1 activation through the released notch
intracellular domain (NICD) and subsequent translocation to the
nucleus. Interaction with TNR induces a repulsion of neurons and
an inhibition of neurite outgrowth (By similarity). {ECO:0000250}.
SUBCELLULAR LOCATION: Isoform 1: Cell membrane; Lipid-anchor, GPI-
anchor; Extracellular side.
SUBCELLULAR LOCATION: Isoform 2: Cell membrane; Lipid-anchor, GPI-
anchor; Extracellular side.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: PG12A_HUMAN FUNCTION: PA2 catalyzes the calcium-dependent hydrolysis of the 2-
acyl groups in 3-sn-phosphoglycerides. Does not exhibit detectable
activity toward sn-2-arachidonoyl- or linoleoyl-
phosphatidylcholine or -phosphatidylethanolamine.
{ECO:0000269|PubMed:12522102}.
SUBCELLULAR LOCATION: Secreted {ECO:0000269|PubMed:12522102}.
Cytoplasm {ECO:0000269|PubMed:12522102}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: LAMC1_HUMAN FUNCTION: Binding to cells via a high affinity receptor, laminin
is thought to mediate the attachment, migration and organization
of cells into tissues during embryonic development by interacting
with other extracellular matrix components.
SUBCELLULAR LOCATION: Secreted, extracellular space, extracellular
matrix, basement membrane.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: CD166_HUMAN FUNCTION: Cell adhesion molecule that mediates both heterotypic
cell-cell contacts via its interaction with CD6, as well as
homotypic cell-cell contacts (PubMed:7760007, PubMed:15496415,
PubMed:15048703, PubMed:16352806, PubMed:23169771,
PubMed:24945728). Promotes T-cell activation and proliferation via
its interactions with CD6 (PubMed:15048703, PubMed:16352806,
PubMed:24945728). Contributes to the formation and maturation of
the immunological synapse via its interactions with CD6
(PubMed:15294938, PubMed:16352806). Mediates homotypic
interactions with cells that express ALCAM (PubMed:15496415,
PubMed:16352806). Required for normal hematopoietic stem cell
engraftment in the bone marrow (PubMed:24740813). Mediates
attachment of dendritic cells onto endothelial cells via homotypic
interaction (PubMed:23169771). Inhibits endothelial cell migration
and promotes endothelial tube formation via homotypic interactions
(PubMed:15496415, PubMed:23169771). Required for normal
organization of the lymph vessel network. Required for normal
hematopoietic stem cell engraftment in the bone marrow. Plays a
role in hematopoiesis; required for normal numbers of
hematopoietic stem cells in bone marrow. Promotes in vitro
osteoblast proliferation and differentiation (By similarity).
Promotes neurite extension, axon growth and axon guidance; axons
grow preferentially on surfaces that contain ALCAM. Mediates
outgrowth and pathfinding for retinal ganglion cell axons (By
similarity). {ECO:0000250|UniProtKB:P42292,
ECO:0000269|PubMed:15048703, ECO:0000269|PubMed:15294938,
ECO:0000269|PubMed:15496415, ECO:0000269|PubMed:16352806,
ECO:0000269|PubMed:24945728, ECO:0000269|PubMed:7760007}.
FUNCTION: Isoform 3: Inhibits activities of membrane-bound
isoforms by competing for the same interaction partners. Inhibits
cell attachment via homotypic interactions. Promotes endothelial
cell migration. Inhibits endothelial cell tube formation.
{ECO:0000269|PubMed:15496415}.
SUBCELLULAR LOCATION: Cell membrane {ECO:0000269|PubMed:15048703,
ECO:0000269|PubMed:15294938, ECO:0000269|PubMed:16352806,
ECO:0000269|PubMed:23169771, ECO:0000269|PubMed:24740813,
ECO:0000269|PubMed:24945728, ECO:0000269|PubMed:7760007}; Single-
pass type I membrane protein {ECO:0000305}. Cell projection, axon
{ECO:0000250|UniProtKB:Q61490}. Cell projection, dendrite
{ECO:0000250|UniProtKB:Q61490}. Note=Detected at the immunological
synapse, i.e, at the contact zone between antigen-presenting
dendritic cells and T-cells (PubMed:15294938, PubMed:16352806).
Colocalizes with CD6 and the TCR/CD3 complex at the immunological
synapse (PubMed:15294938). {ECO:0000269|PubMed:15294938,
ECO:0000269|PubMed:16352806}.
SUBCELLULAR LOCATION: Isoform 3: Secreted
{ECO:0000269|PubMed:15496415}.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS: COX5A_HUMAN FUNCTION: This is the heme A-containing chain of cytochrome c
oxidase, the terminal oxidase in mitochondrial electron transport.
SUBCELLULAR LOCATION: Mitochondrion inner membrane.
CLEAVAGE EVENTS OBSERVED: PLOT OF CLEAVAGE EVENTS:Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' GRP78_HUMAN ELEEIVQPIISK 622 AKKK | ELEE GRP78_HUMAN NDPSVQQDIK 104 GRTW | NDPS GRP78_HUMAN TWNDPSVQQDIK 102 LIGR | TWND GRP78_HUMAN SCVGVFK 40 GTTY | SCVG 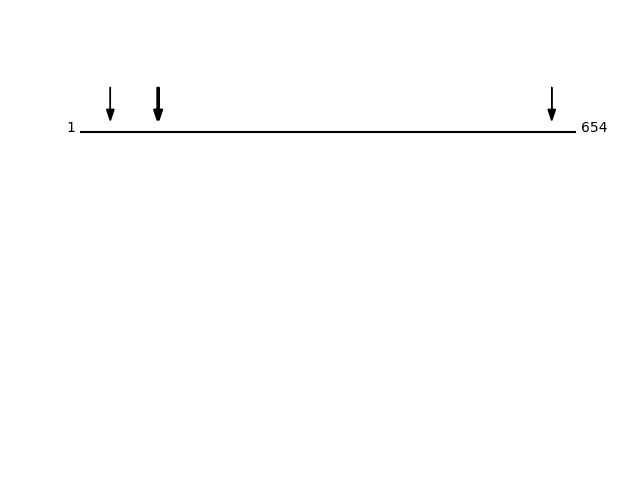
P04843 - Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' RPN1_HUMAN SSEAPPLINEDVKR 25 PGSA | SSEA RPN1_HUMAN SSEAPPLINEDVK 25 PGSA | SSEA RPN1_HUMAN SASSEAPPLINEDVKR 23 PAPG | SASS RPN1_HUMAN SEAPPLINEDVK 26 GSAS | SEAP 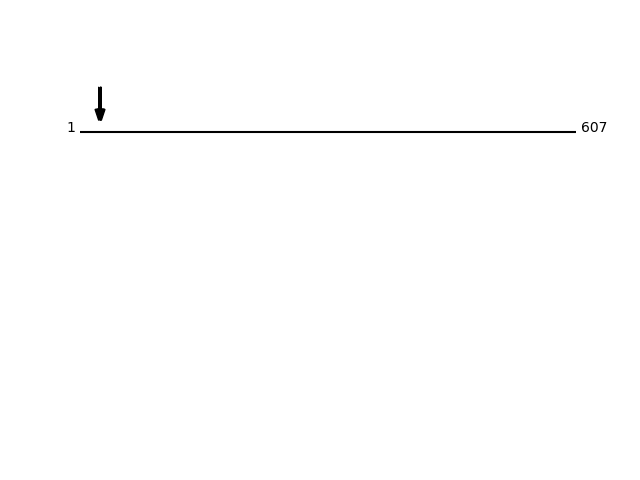
P19022 - Cadherin-2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CADH2_HUMAN TFYGEVPENR 386 FTAM | TFYG CADH2_HUMAN TVAAENQVPLAK 461 MFVL | TVAA CADH2_HUMAN AENQVPLAK 464 LTVA | AENQ CADH2_HUMAN FYGEVPENR 387 TAMT | FYGE CADH2_HUMAN AAENQVPLAK 463 VLTV | AAEN CADH2_HUMAN AFDLPLSPVTIK 638 AGPF | AFDL 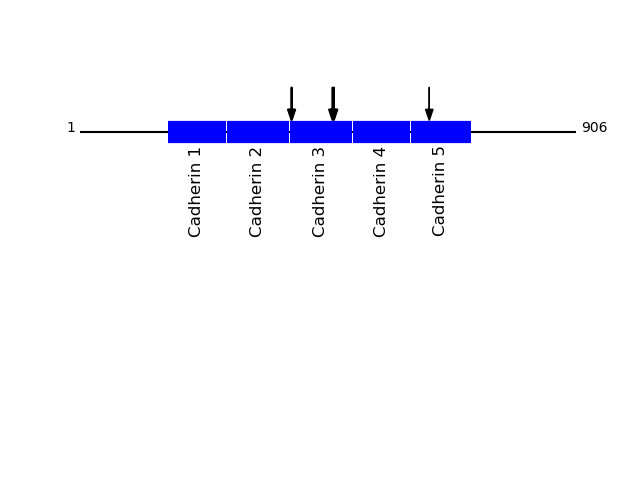
O43852 - CalumeninProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CALU_HUMAN DYDHDAFLGAEEAK 46 AQSF | DYDH CALU_HUMAN TFDQLTPEESK 60 EEAK | TFDQ CALU_HUMAN HLVYESDQNK 272 AEAR | HLVY CALU_HUMAN SHDGNTDEPEWVK 219 GDMY | SHDG CALU_HUMAN ETMEDIDK 195 IVVQ | ETME CALU_HUMAN DQLTPEESK 62 AKTF | DQLT CALU_HUMAN MADKDGDLIATK 162 RRFK | MADK 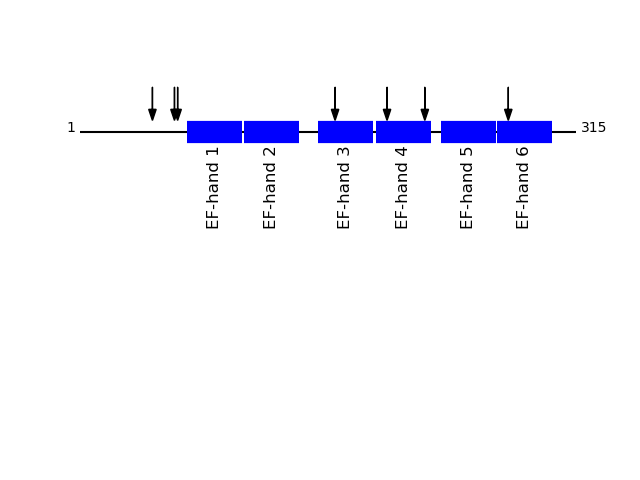
Q5JRA6 - Transport and Golgi organization protein 1 homolog {ECO:0000305}Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TGO1_HUMAN GLAGEPEGELSKEDHENTEK 866 LKTS | GLAG TGO1_HUMAN MAPPLEEGLGGAMEEMQPLHEDNFSR 1043 ATLV | MAPP TGO1_HUMAN EEMQPLHEDNFSR 1056 GGAM | EEMQ TGO1_HUMAN EEMQPLHEDNFSR 1056 GGAM | EEMQ TGO1_HUMAN GLAGEPEGELSK 866 LKTS | GLAG TGO1_HUMAN DVNLQVPDR 755 GRQF | DVNL TGO1_HUMAN EVSQKPNTEK 1097 THAS | EVSQ 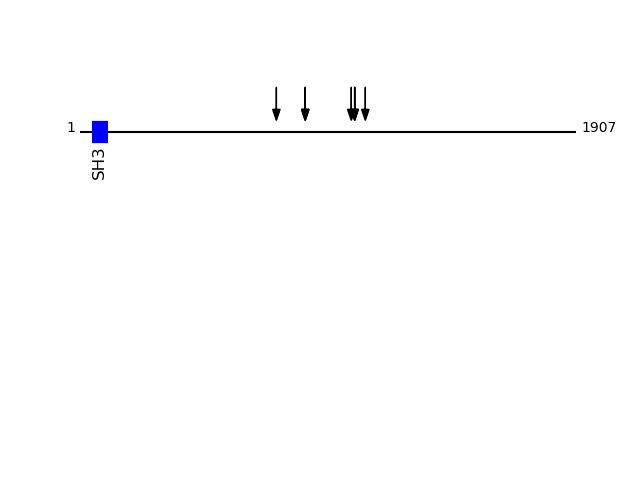
Q12907 - Vesicular integral-membrane protein VIP36Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' LMAN2_HUMAN DITDGNSEHLK 45 SVTA | DITD LMAN2_HUMAN DITDGNSEHLKR 45 SVTA | DITD LMAN2_HUMAN TADITDGNSEHLK 43 LGSV | TADI 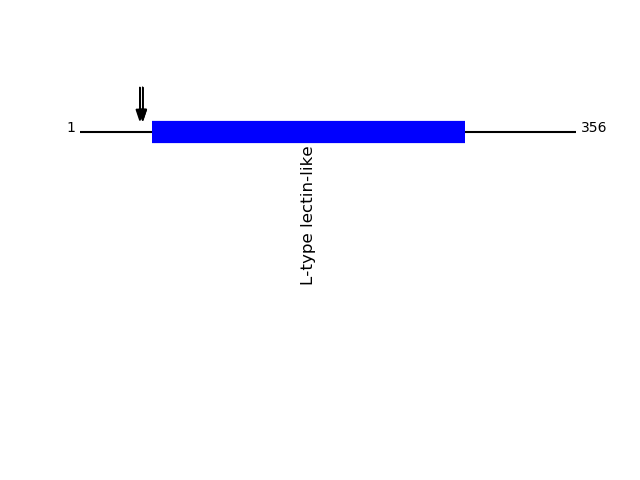
Q8NBS9 - Thioredoxin domain-containing protein 5Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TXND5_HUMAN AAADGPPAADGEDGQDPHSK 44 EAAA | AAAD TXND5_HUMAN AADGPPAADGEDGQDPHSK 45 AAAA | AADG 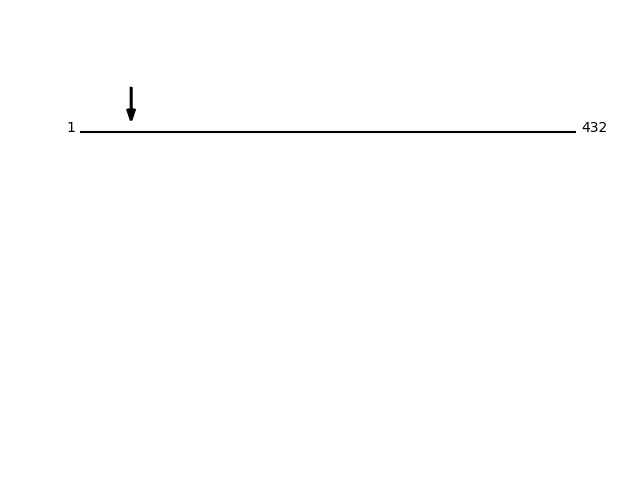
Q99805 - Transmembrane 9 superfamily member 2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TM9S2_HUMAN SVSFEEDDKIR 272 AYTY | SVSF TM9S2_HUMAN DFCQASEGK 82 YTAF | DFCQ TM9S2_HUMAN SFEEDDKIR 274 TYSV | SFEE 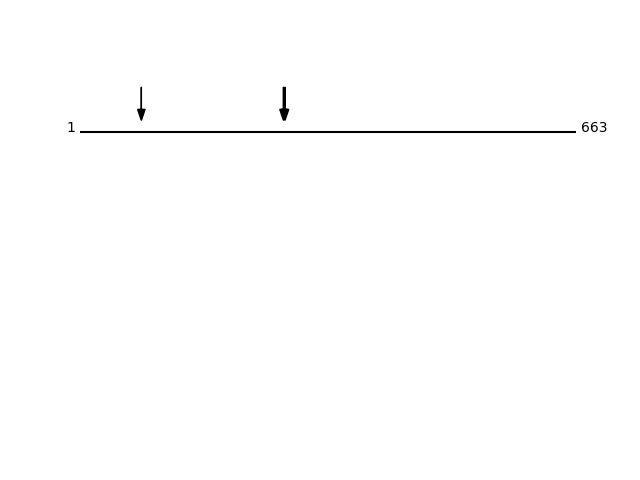
P13667 - Protein disulfide-isomerase A4Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' PDIA4_HUMAN AEGPDEDSSNR 24 AVAG | AEGP PDIA4_HUMAN EGPDEDSSNR 25 VAGA | EGPD PDIA4_HUMAN EVSQPDWTPPPEVTLVLTK 166 AKVR | EVSQ 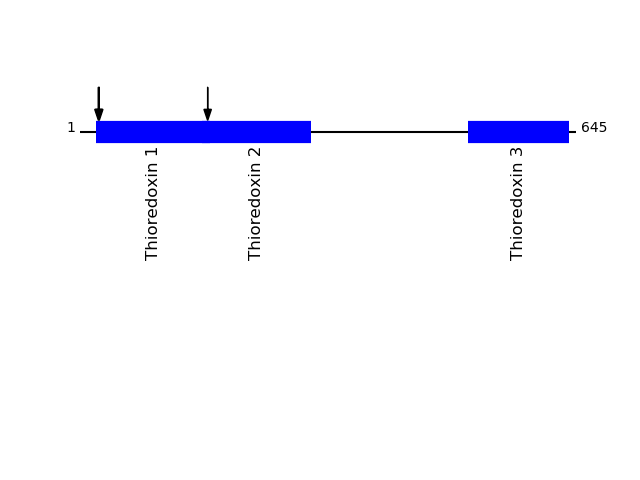
P01889 - HLA class I histocompatibility antigen, B-7 alpha chainProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' 1B07_HUMAN SMYGCDVGPDGR 121 HTLQ | SMYG 1B07_HUMAN GYVDDTQFVR 50 FISV | GYVD 1B07_HUMAN ADTAAQITQR 160 SWTA | ADTA 1B07_HUMAN AAVVVPSGEEQR 269 FQKW | AAVV 1B07_HUMAN VVVPSGEEQR 271 KWAA | VVVP 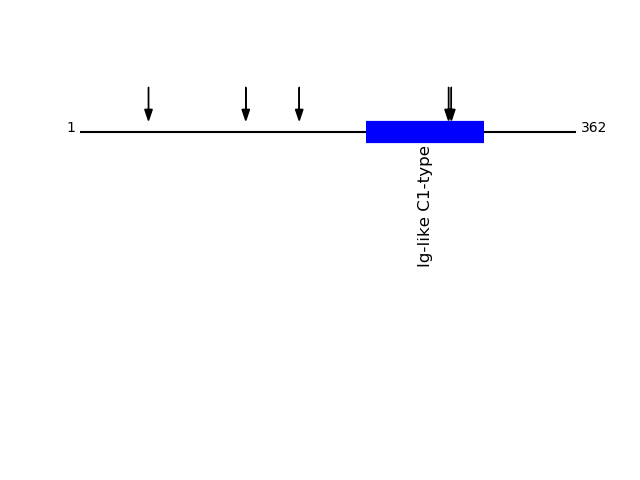
Q14126 - Desmoglein-2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' DSG2_HUMAN DVNDNIPVVENK 264 IRIL | DVND DSG2_HUMAN EVDYEEMK 339 TLIK | EVDY DSG2_HUMAN SLEPAYPPVFYLNK 202 YRIV | SLEP DSG2_HUMAN AWITAPVALR 50 RQKR | AWIT DSG2_HUMAN AISEDYPR 471 VKIV | AISE 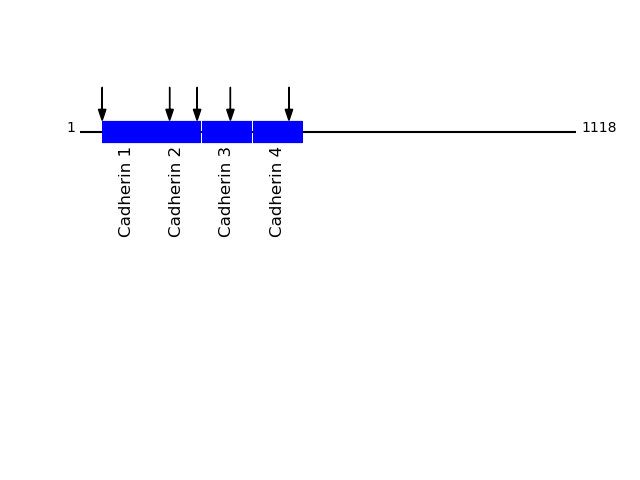
Q9ULF5 - Zinc transporter ZIP10Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' S39AA_HUMAN HEEHDHGPEALHR 26 CNHC | HEEH S39AA_HUMAN GMTELEPSK 42 RQHR | GMTE S39AA_HUMAN GMTELEPSK 42 RQHR | GMTE 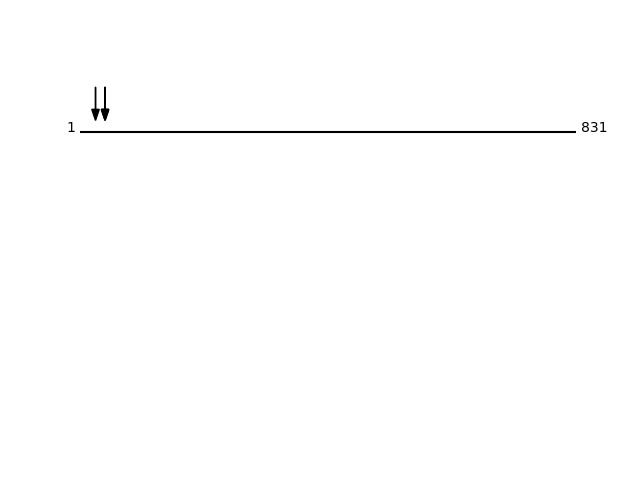
P14314 - Glucosidase 2 subunit betaProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' GLU2B_HUMAN ALLSGDTQTDATSFYDR 255 AEAQ | ALLS GLU2B_HUMAN AQQEQELAADAFK 207 AAAK | AQQE GLU2B_HUMAN SLEDQVEMLR 168 AGKK | SLED 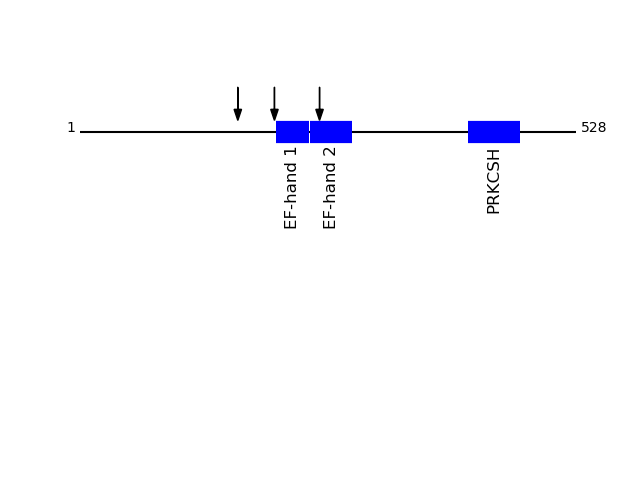
P80303 - Nucleobindin-2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' NUCB2_HUMAN EYHQVIQQMEQK 388 AQKL | EYHQ NUCB2_HUMAN AATSDLEHYDK 161 MLIK | AATS NUCB2_HUMAN EYHQVIQQMEQK 388 AQKL | EYHQ 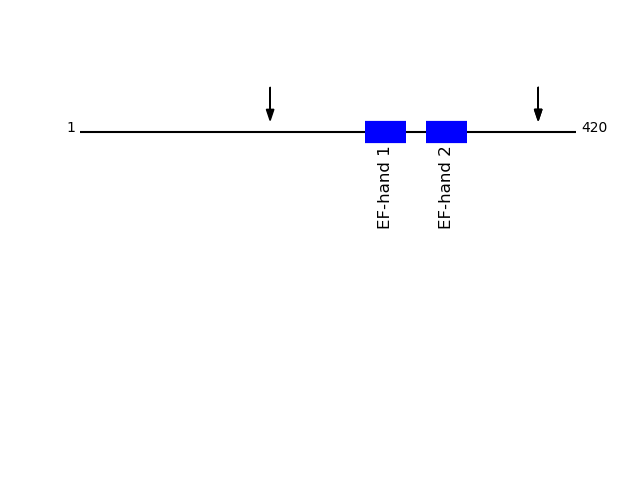
P07237 - Protein disulfide-isomeraseProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' PDIA1_HUMAN DAPEEEDHVLVLR 18 LVRA | DAPE PDIA1_HUMAN NFEDVAFDEK 376 LVGK | NFED 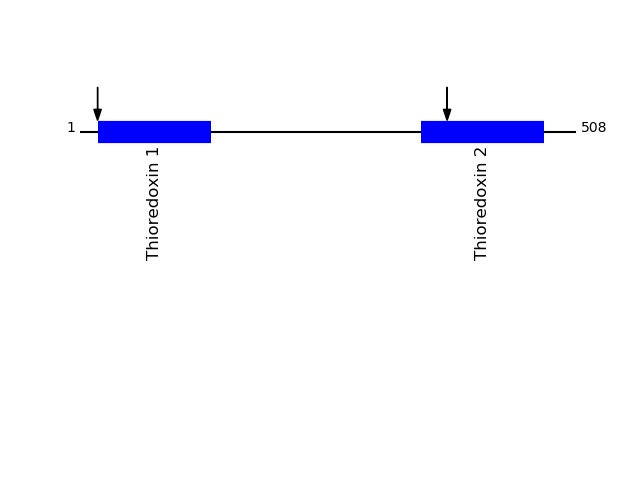
Q15293 - Reticulocalbin-1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' RCN1_HUMAN SHEENGPEPDWVLSER 234 ADMF | SHEE RCN1_HUMAN DLNGDLTATR 179 FKAA | DLNG RCN1_HUMAN EFHDSSDHHTFK 154 GNPA | EFHD RCN1_HUMAN DHEAFLGK 59 SFQY | DHEA 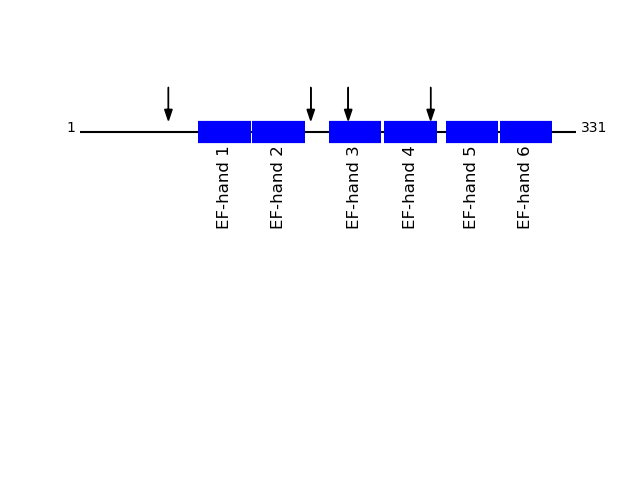
Q14257 - Reticulocalbin-2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' RCN2_HUMAN EALEEHDK 193 FVIQ | EALE RCN2_HUMAN EELHYPLGER 26 AGKA | EELH RCN2_HUMAN ELHYPLGER 27 GKAE | ELHY RCN2_HUMAN DVDEYVK 50 GVQE | DVDE 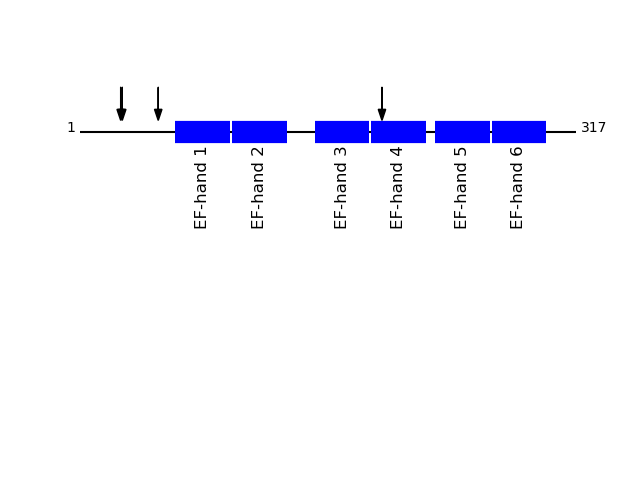
Q9Y624 - Junctional adhesion molecule AProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' JAM1_HUMAN SVTVHSSEPEVR 28 LALG | SVTV 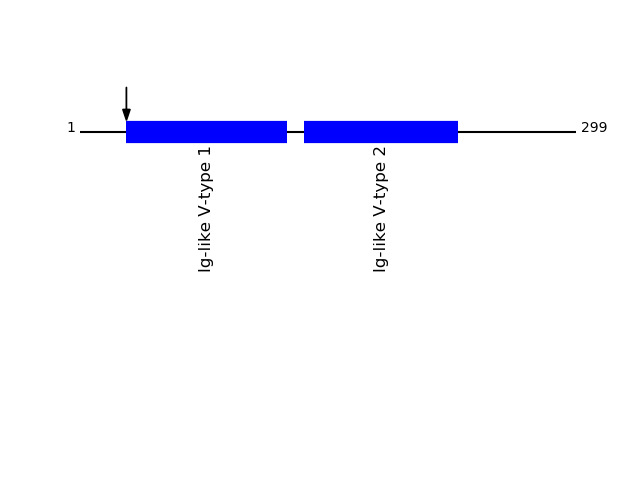
Q9H1E5 - Thioredoxin-related transmembrane protein 4Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TMX4_HUMAN TAGPEEAALPPEQSR 24 AVAA | TAGP TMX4_HUMAN AGPEEAALPPEQSR 25 VAAT | AGPE 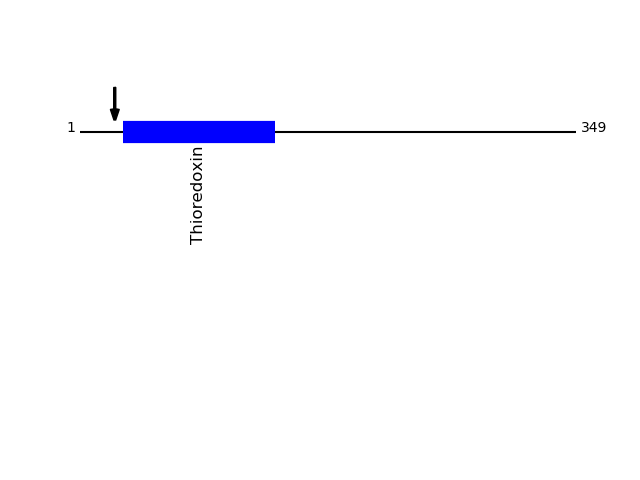
Q04721 - Neurogenic locus notch homolog protein 2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' NOTC2_HUMAN SLPGEQEQEVAGSK 1609 MTRR | SLPG 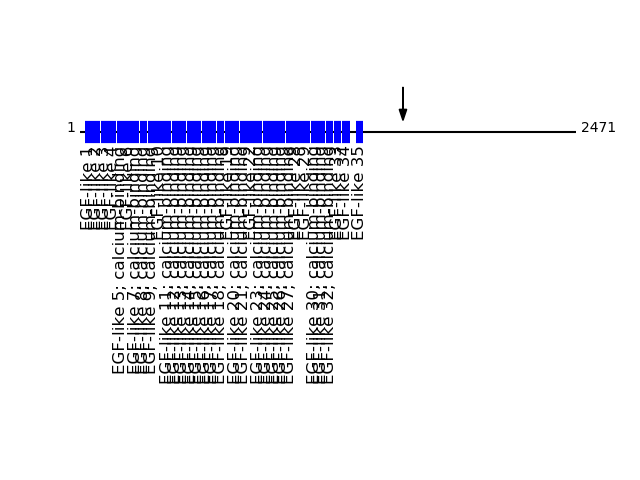
O94901 - SUN domain-containing protein 1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' SUN1_HUMAN AVSEAGASGITEAQAR 608 AVVS | AVSE SUN1_HUMAN AIVNSALK 624 AQAR | AIVN SUN1_HUMAN DAVGQPPR 440 ASVR | DAVG 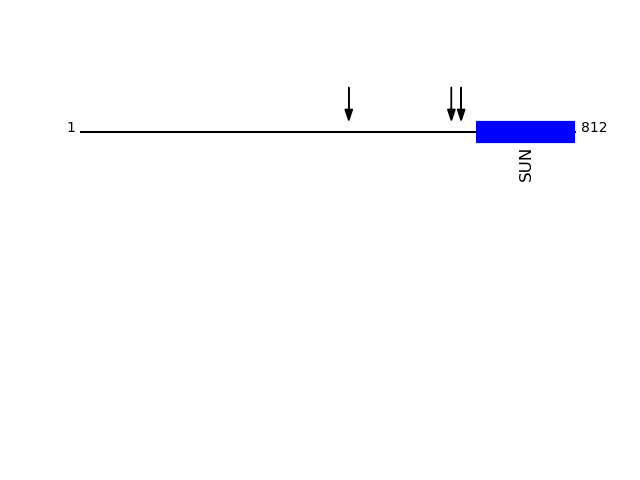
Q92504 - Zinc transporter SLC39A7Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' S39A7_HUMAN GYSHESLYHR 95 GHSH | GYSH S39A7_HUMAN SHESLYHR 97 SHGY | SHES 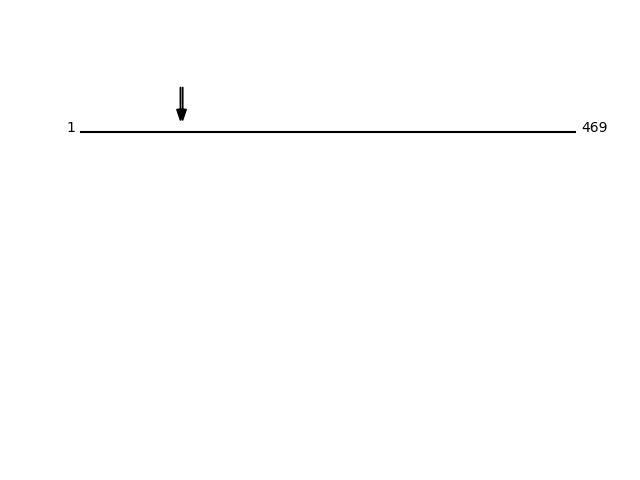
Q14696 - LDLR chaperone MESDProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' MESD_HUMAN AAEGSPGTPDESTPPPR 33 PGSC | AAEG MESD_HUMAN EQWEKDDDIEEGDLPEHK 68 ARLL | EQWE 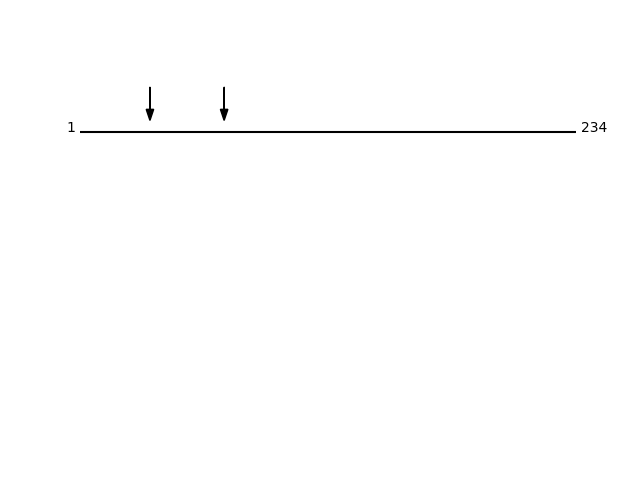
Q8NI22 - Multiple coagulation factor deficiency protein 2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' MCFD2_HUMAN EEPAASFSQPGSMGLDK 27 GARA | EEPA MCFD2_HUMAN SQPGSMGLDK 34 AASF | SQPG 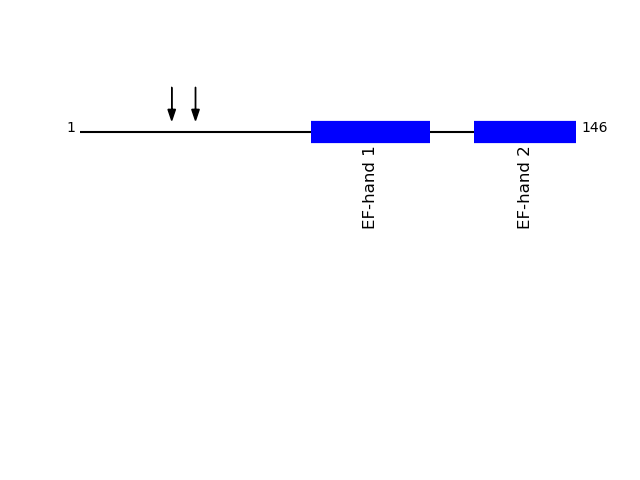
Q9BRN9 - TM2 domain-containing protein 3Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' TM2D3_HUMAN ESTEIPPYVMK 57 PRAA | ESTE TM2D3_HUMAN ESTEIPPYVMK 57 PRAA | ESTE TM2D3_HUMAN EQSQALAQSIK 31 LSGG | EQSQ 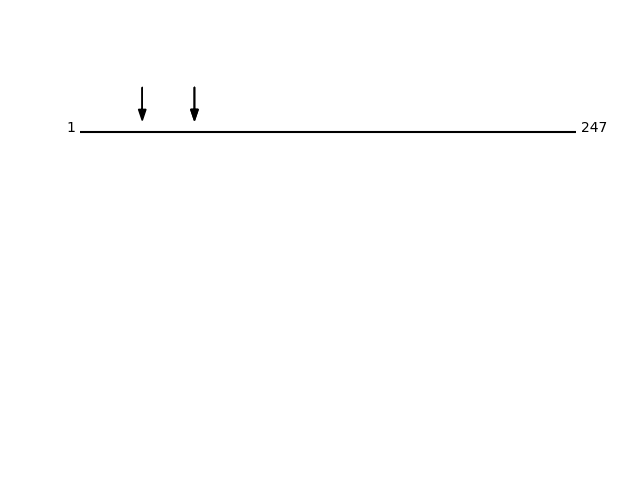
P14209 - CD99 antigenProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CD99_HUMAN SDADLADGVSGGEGK 91 SGSF | SDAD CD99_HUMAN SFSDADLADGVSGGEGK 89 SSSG | SFSD 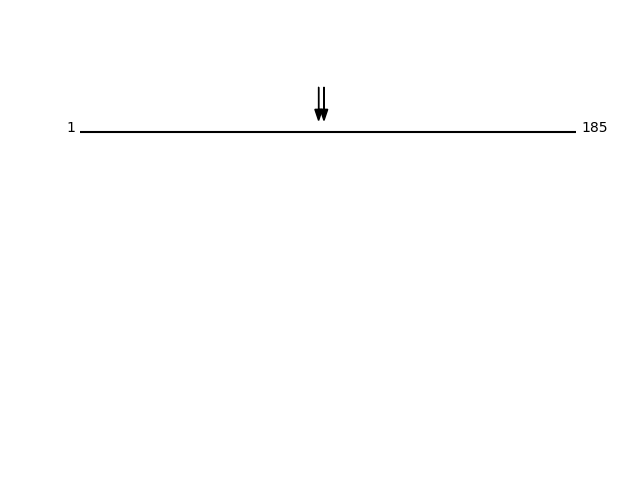
Q9H0V9 - VIP36-like proteinProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' LMA2L_HUMAN GQTFEYLK 45 QVGA | GQTF LMA2L_HUMAN GQTFEYLKR 45 QVGA | GQTF 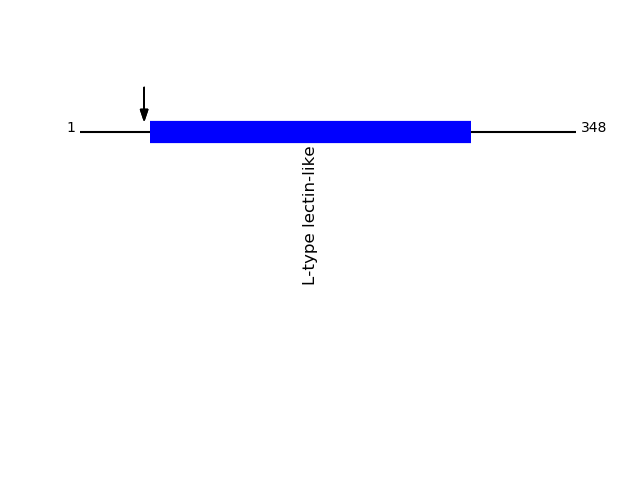
Q8N129 - Protein canopy homolog 4Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CNPY4_HUMAN GMLKEEDDDTER 22 EAWA | GMLK 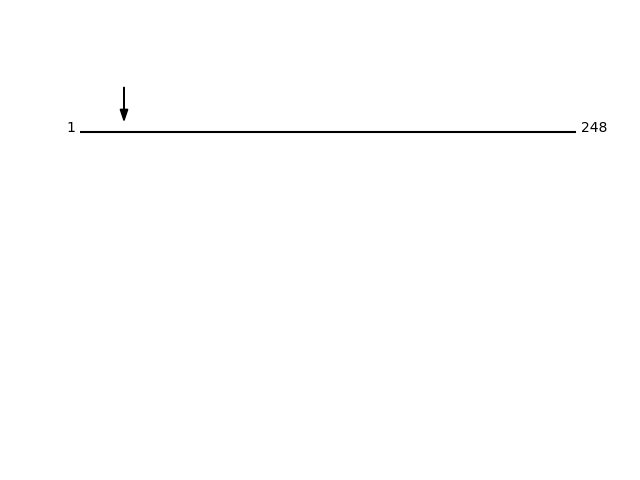
P14625 - EndoplasminProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' ENPL_HUMAN SLNIDPDAK 746 MLRL | SLNI ENPL_HUMAN GWSGNMER 653 ASQY | GWSG 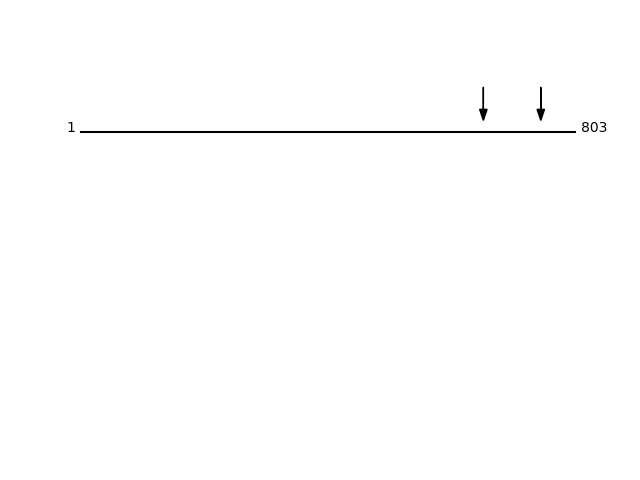
Q07065 - Cytoskeleton-associated protein 4Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CKAP4_HUMAN SLEESEGNKQDLK 275 AKVA | SLEE CKAP4_HUMAN SLEESEGNK 275 AKVA | SLEE 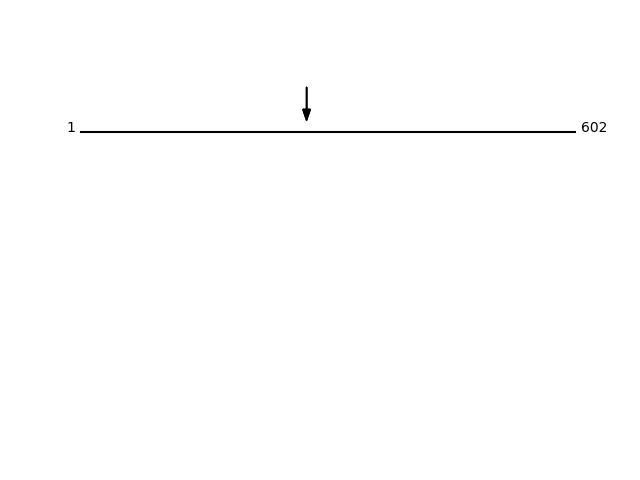
Q13162 - Peroxiredoxin-4Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' PRDX4_HUMAN WETEERPR 38 AVQG | WETE 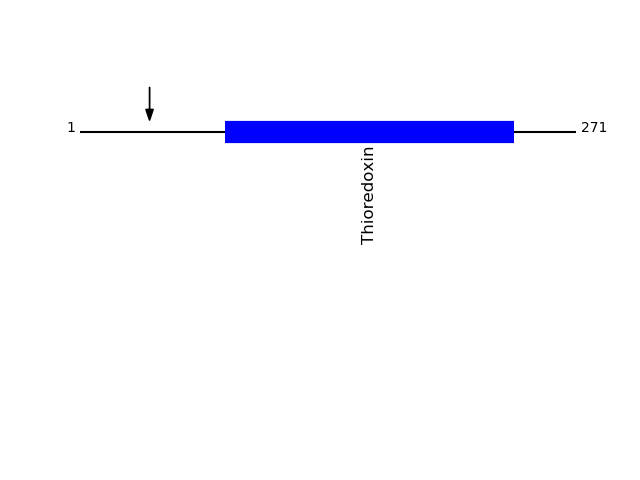
O95980 - Reversion-inducing cysteine-rich protein with Kazal motifsProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' RECK_HUMAN GLAPGSAGALCCNHSK 27 EVAG | GLAP 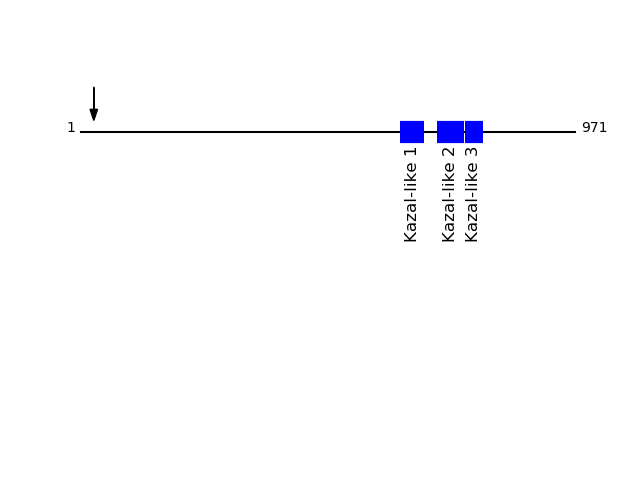
Q12860 - Contactin-1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CNTN1_HUMAN AHSDGGDGVVSQVK 978 VEVR | AHSD CNTN1_HUMAN SAQDAPSEAPTEVGVK 803 AVIN | SAQD 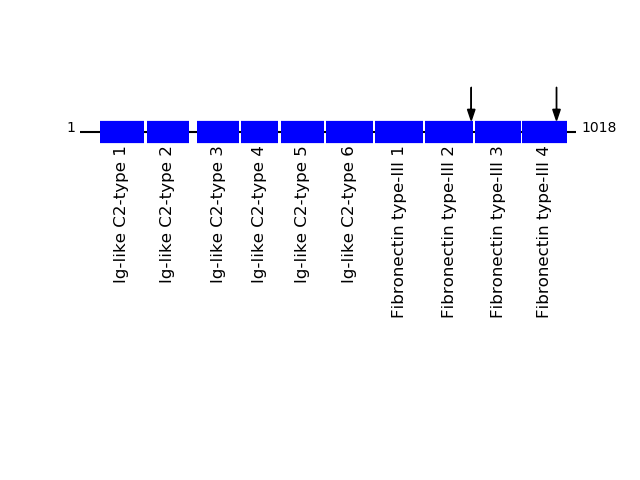
Q9BZM1 - Group XIIA secretory phospholipase A2Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' PG12A_HUMAN ALDLLGGEDGLCQYK 52 YLNA | ALDL 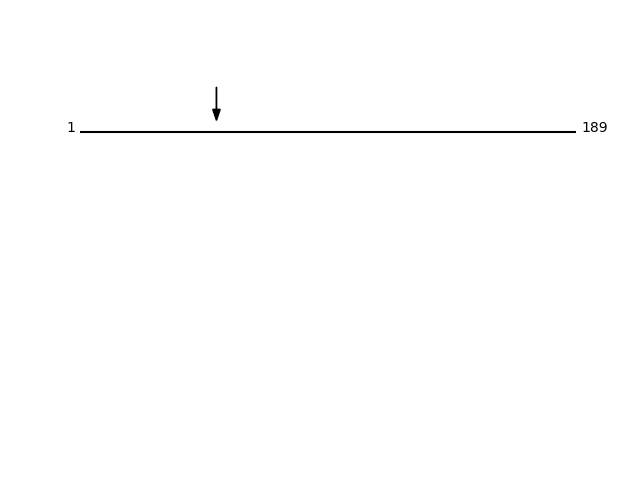
P11047 - Laminin subunit gamma-1Protein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' LAMC1_HUMAN SQAAQEAEINAR 1493 AGMA | SQAA LAMC1_HUMAN ASQAAQEAEINAR 1492 MAGM | ASQA 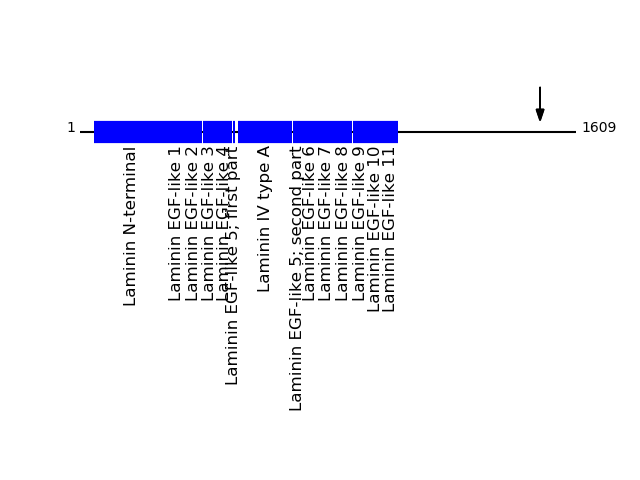
Q13740 - CD166 antigenProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' CD166_HUMAN SIPEHDEADEISDENR 504 VSAI | SIPE 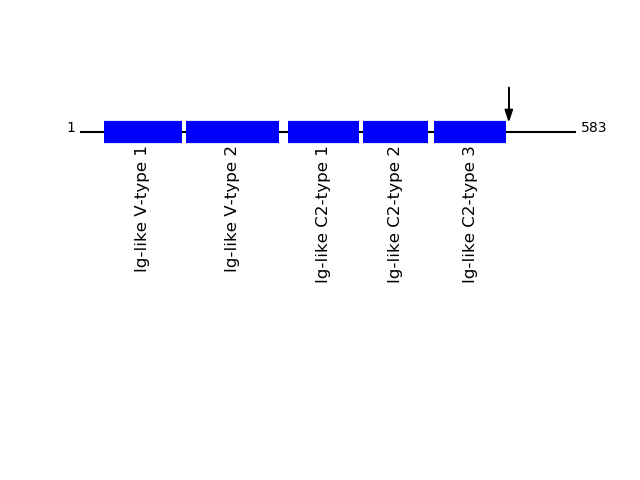
P20674 - Cytochrome c oxidase subunit 5A, mitochondrialProtein Peptide Cleavage Position (P1') Cleavage Site P4-P1 | P1'-P4' COX5A_HUMAN SHGSQETDEEFDAR 42 VRCY | SHGS